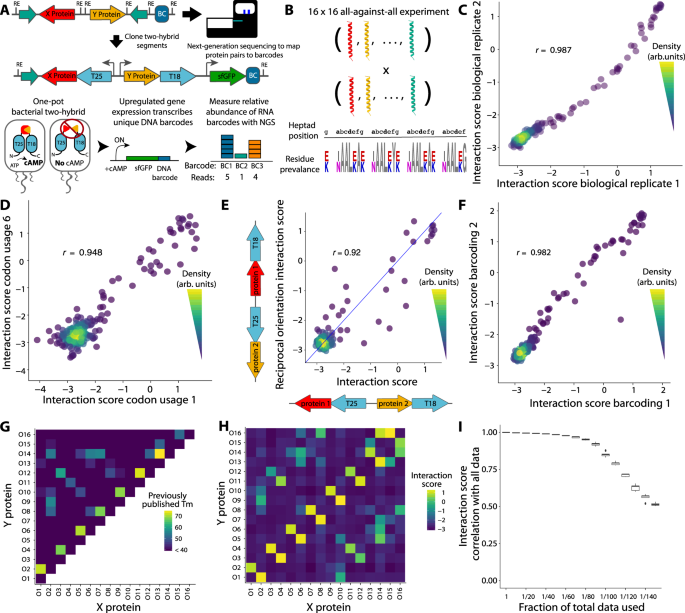
NGB2H system design
Regardless of a wealth of methods by way of which to analyse PPIs, there may be not at the moment a way that facilitates high-throughput characterisation when analysing PPIs in codecs aside from all-against-all or is ready to distinguish between carefully associated constructs. Nonetheless, such a system would enable investigations of PPIs inside protein households, polymorphic PPIs, and de novo designed PPIs which can be at the moment intractable. Thus, we constructed a generalisable, scalable bacterial two-hybrid system utilizing a considerably modified model of the B. pertussis adenylate cyclase two-hybrid17 (Fig. 1A, Supplementary Data Part 1). Briefly, the two-hybrid features a lot as in Karimova et al.17, through which interacting hybrid proteins reconstitute adenylate cyclase to supply cAMP, which drives reporter gene expression. We measured the relative transcription of a uniquely figuring out DNA barcode residing within the reporter gene, which serves as a measure of interplay energy. The barcode is mapped to the 2 absolutely sequenced hybrid proteins at an early cloning step utilizing high-throughput sequencing when the barcode and proteins are bodily adjoining. This unambiguously identifies even extremely homologous proteins and separates artificial errors from programmed designs. Thus, measuring the relative barcode transcription supplies a quantitative, massively multiplexed characterization of PPIs with short-read sequencing. As a result of the NGB2H system makes use of a mapping step, it will possibly use gene synthesis, somewhat than preconstructed libraries, to create range, which additional frees it from the one-against-all or all-against-all testing frequent in two-hybrids. We made numerous different enhancements, together with (1) titratable and inducible management of hybrid protein expression and optimised reporter response on a single plasmid, (2) a background pressure with linear cAMP accumulation, (3) a inexperienced fluorescence protein (GFP) reporter as a substitute of beta-galactosidase for extra fast particular person characterisation, (4) using a number of barcodes per assemble to attain statistically strong outcomes and (5) a scarless cloning scheme that enables for library creation with any designed sequence (extra data in Supplementary Data Part 1).
A High: schematic of the subsequent technology bacterial two-hybrid (NGB2H) system cloning and assemble. T25, T18 – adenylate cyclase halves; BC – distinctive DNA barcode figuring out the protein pair. Backside: Workflow of NGB2H system. Interacting proteins reconstitute adenylate cyclase, producing cAMP (cyclic adenosine monophosphate), which drives the gene expression of the barcoded super-folder inexperienced fluorescence protein (sfGFP) reporter. Relative barcode abundance is quantified utilizing next-generation sequencing (NGS). B The CC0 Library consists of 16 coiled-coils examined towards each other. (Backside) Sequence brand representing the variety within the CC0 Library. Residues that fluctuate are proven in color. C Interplay scores of CC0 library members are constant between organic replicates (Pearson’s r > 0.98). D Two totally different codon usages have constant interplay scores (Pearson’s r > 0.94, consultant pattern). E Interplay energy is analogous (Pearson’s r > 0.92) no matter which protein is connected to which half of adenylate cyclase. The blue line represents y = x. F Interplay scores of individually barcoded, cloned, and examined replicates are constant (Pearson’s r > 0.98). G Printed round dichroism (CD) melting level (Tm) information. H Experimentally decided interplay scores. I CC0 library Uncooked information might be subsampled and nonetheless correlate effectively with the total dataset. Boxplot heart traces signify the median, the hinges signify the twenty fifth and seventy fifth percentiles and whiskers signify the most important/smallest worth inside 1.5x it’s respective hinge for 50 subsamples with substitute of the total information. Supply information are supplied as a Supply Knowledge file.
Validation of the NGB2H system
After optimising the system with single-construct GFP measurements (Supplementary Fig. 1), we validated the NGB2H system with 256 beforehand characterised interactions15, which we name the CC0 Library. The CC0 Library is a set of sixteen de novo designed, orthogonal, heterodimeric coiled-coils which can be examined in an all-against-all configuration. The proteins are extremely related, being 4 heptad coiled-coils that fluctuate solely on the a-position (Ile/Asn), e-position and g-position (Lys/Glu) (Fig. 1B). We designed the CC0 Library to be suitable with our system (Supplementary Fig. 2A) after which barcoded and cloned it (Supplementary Figs. 3A, 4). After inducing the two-hybrid for six hours, we took samples for RNA and DNA extraction to measure the interplay energy and normalize for plasmid abundance, respectively. We obtained high-quality measurements for all 256 protein pairs and calculated an interplay rating, outlined because the pure logarithm of the median of the ratio of the RNA to DNA reads: ({Interacti}{on; rating}={{{{{rm{ln}}}}}}left({{{{{rm{median}}}}}}left(frac{{RNA; reads}}{{DNA; reads}}proper)proper)).
Solely barcodes for which ten or extra reads had been obtained in each DNA replicate and that completely mapped to designed protein pairs had been utilized in additional evaluation. The NGB2H assay was extremely replicable, with organic replicates having related interplay scores (Pearson’s r > 0.98, p < 10−15), with a dynamic vary of greater than 100-fold (Fig. 1C).
We checked a number of inside controls to validate the measurements of the NGB2H assay. First, as a result of the protein code is degenerate, we screened 9 codon usages for every pair of proteins. Completely different codon usages confirmed constant interplay scores (consultant pair Fig. 1D), with all usages correlating with Pearson’s r > 0.92 and p < 10−15 (Supplementary Fig. 5), demonstrating minimal results on the a part of DNA sequence variation and low ranges of noise within the interplay scores. We additionally in contrast the interplay scores of protein pairs when the 2 constituent proteins had been connected to the opposite half of the two-hybrid, which we name the reciprocal orientation. We discovered that the CC0 Library reveals a robust correlation between the first and reciprocal orientations (Pearson’s r = 0.92, p < 10−15, Fig. 1E), indicating that the organic equipment of the NGB2H system faithfully recapitulates the biochemical interplay. As well as, a portion of our library contained frameshift mutations, which mustn’t create purposeful PPIs. As anticipated, the interplay scores of constructs with indels are clustered on the backside of the vary of appropriate constructs (Supplementary Fig. 6). Final, to point out that the NGB2H system doesn’t endure from barcode results or choice stress from the repeated cloning steps, we replicated the assay with an unbiased re-barcoding and re-cloning of the CC0 Library, which confirmed robust correlation with the primary iteration’s interplay scores (Pearson’s r > 0.98, p < 10−15, Fig. 1F).
Having confirmed the inner consistency of the CC0 Library, we in contrast it to the beforehand revealed outcomes. In comparison with the round dichroism information revealed in Crooks et al.15, we discovered that the NGB2H system’s dynamic vary correlated effectively with melting temperatures larger than 40 °C (Fig. 1G, H). Given the variations in method – in vivo versus in vitro, interplay energy versus helicity – the correlation between the interplay rating and melting level temperatures (Pearson’s r > 0.75, p < 10−15, Supplementary Fig. 7) largely validate the NGB2H system. Lastly, the NGB2H system have to be extremely scalable. To check its scalability, we computationally lowered the variety of reads used within the evaluation between 10 and 150-fold and located robust settlement with our full dataset, even when the uncooked information had been lowered 100-fold (Pearson’s r > 0.85, p < 10−15, Fig. 1I), which means the flexibility to precisely display screen ~25,000 interactions at an analogous learn depth.
Design of enormous units of orthogonal coiled-coils
All dimeric coiled-coils have an analogous construction, which is why sequence-based scoring features can fruitfully predict melting temperatures or binding affinities. The scoring features settle for two sequences as enter, normally starting with a particular register, and return a rating. One of many broadly used algorithms is bCipa14, which is predicated on summing weights for residue-residue interplay pairs, in addition to electrostatic interactions and helical propensity, and predicts melting temperatures. The state-of-the-art scoring perform was developed by Potapov et al.13, which makes use of triplet weights, along with the pair weights, and a a lot bigger coaching set to foretell the free power of binding. The paper additionally benchmarks the most typical CC scoring features, resembling Fong/SVM18 and Vinson/CE12.
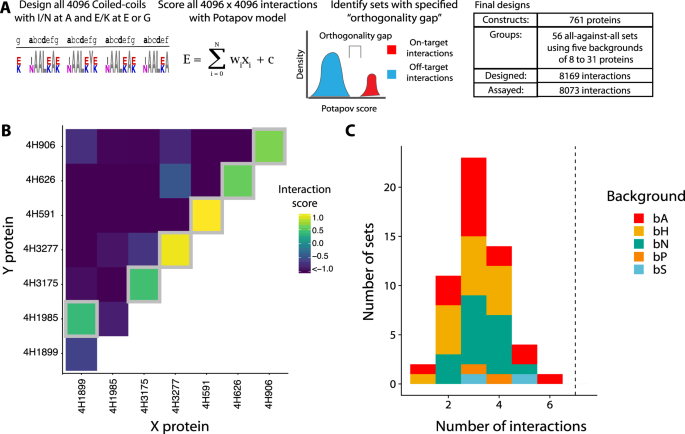
To computationally predict massive, orthogonal units of coiled-coils for empirical verification, we constructed a two-step computational pipeline (Fig. 2A). In short, we calculated 16.7 million scores for all dimeric interactions between four-heptad coiled-coils with Ile or Asn on the a-position and Glu or Lys on the e– and g– positions utilizing the scoring mannequin of Potapov et al.13. The floor b-, c– and f-positions had been set to Ala. We then recognized orthogonal units, which might be divided into on-target and off-target interactions such that every constituent protein participates in precisely one on-target interplay, which is stronger than each off-target interplay. This enables us to outline an orthogonality hole for an orthogonal set, the place the orthogonality hole is calculated because the weakest on-target interplay minus the strongest off-target interplay. For instance, in Fig. 2B, on-target interactions are on the diagonal (homodimers) or simply above the diagonal (heterodimers). All different interactions are thought-about off-target. Although computationally difficult, figuring out units with an orthogonality hole is tractable as a variant of the utmost unbiased set downside19. Utilizing the bCipa and Potapov scoring features, we recognized the fifteen largest units and included every of them with three totally different units of residues on the b-, c– and f-positions as a result of floor positions can modulate dimer stability and solubility20. We discuss with a set of residues used on the b-, c– and f– positions as backgrounds as a result of these don’t have an effect on orthogonality. We mixed these with two units of controls spanning eleven backgrounds, leading to a complete of 56 units containing between 64 to 961 interactions (8169 interactions general), which we named the CCNG1 Library. After testing a subset of the CCNG1 Library to validate our in-house designs, which we name the CC1 Library, (see Supplementary Figs. 8, 9; Supplementary Data Part 8.3), we designed (Supplementary Fig. 2C), cloned (Supplementary Fig. 3C, 4), and carried out the NGB2H assay, from which we collected high quality information (Supplementary Fig. 10) on 8073 interactions. The CC0 Library was added to the CCNG1 library as an inside management (Supplementary Fig. 10C).
A Schematic of the CCNG1 Library design. All four-heptad coiled-coils with variation on the a-, e-, and g– positions had been scored for interactions with the mannequin of Potapov et al., and subsets of coiled-coils with massive orthogonality gaps had been recognized. In whole, we designed and examined 56 units of orthogonal coiled-coils. B The orthogonal subset of coiled-coils with the most important variety of on-target interactions (six on-target interactions). Gray bins determine on-target interactions. C Variety of interactions per orthogonal subset of coiled-coils. Dashed line represents the variety of on-target orthogonal interactions within the CC0 library. Colors present the totally different backgrounds used, whereas the interfacial residues remained the identical. Supply information are supplied as a Supply Knowledge file.
The area of all doable pairs, assuming solely our restricted set of amino acid residues (~16 M), is a number of orders of magnitude bigger than what could possibly be screened experimentally (~25 ok), so the design course of is essential in figuring out possible orthogonal units that may be experimentally examined.
Massive orthogonal units within the CCNG1 library
Though we designed our coiled-coils to type orthogonal units, the present state-of-the-art coiled coil scoring features usually are not sufficiently correct to take action reliably, and practically all units contained off-target interactions stronger than among the on-target pairs. The proteins concerned in robust off-target interactions might be faraway from the set, leaving solely these interactions which can be experimentally verified to be orthogonal. Thus, we discuss with an orthogonal subset as the most important experimentally characterised group of orthogonal interactions amongst what was computationally predicted to be an orthogonal set. To determine the orthogonal subset of every designed orthogonal set, we used an analogous method to that described above and lowered the issue to the utmost unbiased set downside utilizing Interplay scores from the NGB2H assay.
To make our outcomes strong to experimental noise from the NGB2H assay, we would have liked to search out an applicable orthogonality hole, that’s bigger than the uncertainty of the interactions rating. We now have carried out a radical evaluation of each the CC0 inside management (technical repeats), exterior controls (comparability to measured melting factors, Supplementary Fig. 10C) and particularly the supply of reciprocal enzyme orientations for a similar peptide pair (pairs of equivalent peptides the place the break up cAMP components are reversed). We discovered uncertainty of lower than 0.8 interplay scores in all experiments on this paper (Supplementary Knowledge 9). Thus, to be conservative we enforced an orthogonality hole of at the very least 1.0 Interplay Rating. Utilizing this framework, we had been in a position to determine an orthogonal subset of coiled-coils that incorporates six pairs, which incorporates one heterodimer and 5 homodimers (Fig. 2B). The orthogonality hole we implement could be very strict, for instance the CC0 management set has a spot of solely 0.4, and on the orthogonality hole of 1.0 it incorporates solely 4 pairs as a substitute of seven.
There are additionally functions the place the necessities for orthogonality might be lowered, for instance in constructing protein origami as demonstrated by Aupič et al.21, through which two equivalent pairs had been utilized in the identical construction. Pairwise orthogonality is probably the most stringent criterion. In a single-pot experiment, through which all pairs could be current, we speculate that orthogonality would solely enhance as a result of the off-target states could be competing with the on-target states.
Subsequently we have now additionally calculated orthogonal units with orthogonality gaps of 0.0 and 0.5. At an orthogonality hole of zero, 20 of our 51 experimentally recognized orthogonal subsets in CCNG1 library had greater than the seven on-target orthogonal interactions (Supplementary Fig. 13). Orthogonal units at totally different orthogonality gaps are introduced in Supplementary Knowledge 6.
The CCNG1 Library represents the primary large-scale systematic investigation of the consequences of variation on the b-, c-, and f-positions; subsequently, we sought to grasp how these positions influenced interactions. As anticipated, we discovered that totally different backgrounds didn’t considerably have an effect on orthogonality (Fig. 2C and Supplementary Figs. 11, 12). We examined six backgrounds containing the identical interfacial residues because the CC0 Library (Supplementary Fig. 14 and Supplementary Data Part 8.4) and located that charged however much less helical backgrounds led to weaker, much less particular interplay profiles. The findings agree with the mannequin introduced by Drobnak et al.22, through which the b-, c– and f– positions had been used to modulate affinity.
Enchancment of coil-coiled interaction-prediction algorithms
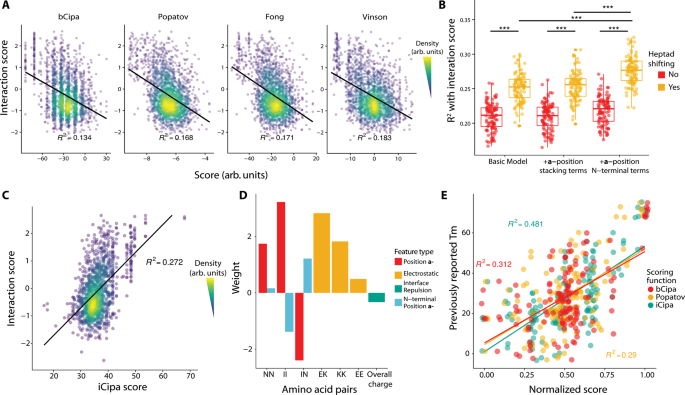
The CCNG1 Library dataset represents the most important dataset of coiled-coil interactions so far. We reasoned that our information may function a coaching set to enhance on at the moment out there fashions. To benchmark present fashions, we computed scores utilizing the algorithms bCipa14, Potapov/SVR13, Fong/SVM18 and Vinson/CE12, that are all linear fashions with options for amino acid pairings. Every algorithm is barely weakly predictive of our measured interactions with the bA background (Fig. 3A) as a result of all fashions have an R2 < 0.2. Notably, every algorithm predicted the strongest interactions effectively but in addition predicted many weak interactions that, when measured, had excessive interplay scores.
A Earlier fashions of coiled-coils predict the interactions within the CCNG1 with low R2. The black line represents a linear mannequin of the interplay scores predicted by totally different algorithms. B Coefficient of willpower of interplay scores of various iCipa candidates evaluated throughout improvement. Every level represents one bootstrap of the info. N = 100 bootstraps. Boxplot heart traces signify the median, the hinges signify the twenty fifth and seventy fifth percentiles and whiskers signify the most important/smallest worth inside 1.5x it’s respective hinge. ***p < 10−15 by two-tailed t-test. C iCipa is extra predictive of interplay scores (R2 > 0.27) than the earlier fashions proven in (A). Black line represents a linear mannequin of interactions scores, as predicted by iCipa scores. D Weights for the iCipa mannequin. Every weight scores a pair of amino acid residues at particular registers between the coiled coils (at a–a’: NN and II, at e–g’ and g–e: KE, KK and EE). E iCipa is extra predictive of the beforehand revealed CC0 melting factors than the bCipa or Potapov scoring features. Particular person dots signify melting factors as in contrast with the normalised rating from one of many three scoring algorithms. Boxplot heart traces signify the median, the hinges signify the twenty fifth and seventy fifth percentiles and whiskers signify the most important/smallest worth inside 1.5x it’s respective hinge. Supply information are supplied as a Supply Knowledge file.
We constructed a number of linear fashions much like bCipa, which included quite a few improvements (Supplementary Data Part 3). First, we educated a mannequin on our information that solely included weights for the a-, d-, e– and g– place mixtures. We additionally created variations of this easy mannequin with phrases for both consecutive residues within the a- place of the identical protein or separate phrases for weights on the N-terminal a– place, the place fraying could happen (Supplementary Fig. 15A).
We then expanded these fashions with a scoring method, which we name heptad shifts (Supplementary Fig. 15B). Briefly, we count on the predominant type of coiled-coil interplay to be the alignment of heptads which have the strongest interplay. By way of the massive variety of off-target interactions, this doesn’t essentially point out that each one 4 heptads are aligned with the N-terminus however, somewhat, may point out an interface of three or fewer heptads. We now have educated the fashions iteratively by altering the alignment of off-target pairs, retraining the fashions and rescoring the off-target alignments till convergence was achieved (in lower than 5 repetitions in all circumstances). All of our heptad-shifting scoring algorithms had been considerably higher than the corresponding non-shifting variations. Our N-terminal a– place weights algorithm was considerably higher than each the fundamental algorithm and the consecutive a– place algorithm (Fig. 3B). Thus, our last mannequin, which we name iCipa, makes use of heptad shifting and phrases for the N-terminal a– positions, and it’s extra predictive of CCNG1 Interplay scores than earlier fashions, with an R2 = 0.27 (Fig. 3C). The impact of heptad shifting on iCipa, in addition to bCipa and the Potapov scoring perform, is proven in Supplementary Fig. 16.
iCipa is a linear mannequin, which facilitates interpretation. The weights of iCipa have anticipated and sudden traits (Fig. 3D). a– place residues favor Ile/Ile pairings, tolerate Asn/Asn pairings between proteins and disfavour Ile/Asn pairings, as anticipated. As anticipated, the e– and g– positions favour salt bridges between Glu/Lys and disfavour Glu/Glu pairings. Maybe counterintuitively, Lys/Lys pairings are acceptable, and former biochemical work has recognized mildly beneficial binding contributions on the a part of Lys/Lys pairings23.
To check the iCipa mannequin, we excluded all the info from the unique CC0 Library whereas we educated the weights. When the scoring features are normalised and in contrast (Fig. 3E), each the Potapov/SVR and bCipa algorithms carried out worse when it comes to predicting the measured melting temperatures, with R2 < 0.32, as in comparison with iCipa, with R2 = 0.48, representing a 50% enhance in predictive capacity. Importantly, the rise in predictive energy for iCipa on the CC0 Library demonstrates that iCipa has not been educated on an artifact of the NGB2H system however, somewhat, that the NGB2H system supplies high-quality information on PPIs, which may present basic insights into coiled-coil perform.
CCmax library design and verification
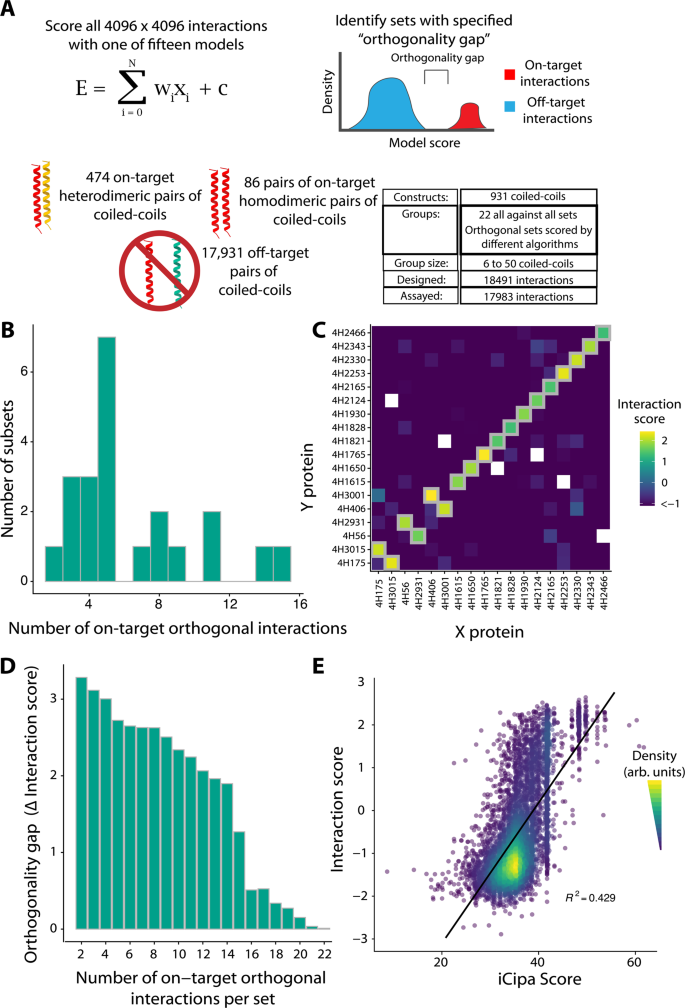
To judge iCipa’s prediction capabilities, reveal the scalability of the NGB2H system, and determine bigger orthogonal units of coiled-coils, we constructed one other library, the CCmax Library. The CCmax Library incorporates 18,491 interactions and incorporates 931 totally different coiled-coils in fifteen predicted orthogonal units and 7 management units (Fig. 4A). The orthogonal units had been designed utilizing our computational framework and scored with one in every of fifteen variants of iCipa. After designing (Supplementary Fig. 2D) and cloning, we collected high-quality information on 17,983 interactions (Supplementary Fig. 17). The CC0 Library was an inside management added to the CCmax Library, and it broadly agreed with its efficiency in our earlier libraries (Supplementary Fig. 18).
A Design of the CCmax library. Utilizing a number of iCipa variants 22 units comprised of 18,491 interactions had been designed and orthogonal units of orthogonal interactions with a given orthogonality hole had been recognized (Supplementary Knowledge 6). B Variety of on-target orthogonal interactions per orthogonal subset at orthogonality hole of 1.0 Interplay Rating. Between two and fifteen on-target, orthogonal interactions had been obtained per subset. C The most important orthogonal subset incorporates fifteen on-target interactions throughout 318 examined pairs. Gray bins signify designed on-target interactions. D The most important orthogonality hole per quantity on-target interactions in a set. E iCipa’s settlement with the Interplay rating (R2 = 0.429). The black line is a linear mannequin predicting interplay scores from iCipa predictions. Supply information are supplied as a supply information file.
Orthogonal units of the CCmax library
Equally to the CCNG1 library, we recognized the most important experimentally recognized orthogonal subsets of every designed set with an orthogonality hole of 1.0 Interplay Rating. These orthogonal subsets have as many as fifteen on-target pairs (Fig. 4B) and 318 whole interactions from 18 totally different proteins (Supplementary Fig. 19). 5 of the orthogonal subsets contained extra on-target interactions than the most important revealed coiled coil set15. Our largest orthogonal subset (Fig. 4C) contained fifteen coiled-coil dimers, twelve homodimers and three heterodimers, which is 9 extra on-target interactions than the set from CCNG1, displaying the advance of iCipa over bCipa and the Potapov scoring features.
Just like the CCNG1 Library, we additionally recognized units with decrease orthogonality gaps of at the very least 0.0 Interplay Rating, 0.5 Interplay Rating, and one RMSD between the reported melting temperatures of the CC0 subset of the CCmax library mapped to Interplay Scores (Supplementary Fig. 17C). Reducing the orthogonality hole recognized extra interactions with a most of twenty-two on course interactions from twenty-eight totally different proteins when the hole is zero (Supplementary Fig. 20). All of the orthogonal units are listed in Supplementary Knowledge 6.
Completely different functions require totally different ranges of orthogonality; whereas gene circuits doubtless require excessive orthogonality, protein origami, which advantages from avidity, just isn’t underneath such strict constraints. Thus, we recognized the most important orthogonality hole for various numbers of on-target interactions (Fig. 4D; Supplementary Knowledge 7). As anticipated, smaller units had bigger gaps, however orthogonality gaps of at the very least 0.5 interplay Rating had been recognized for units as massive as seventeen on-target interactions. Lastly, we in contrast the CCmax Library’s interplay rating with the iCipa predictions, which present substantial enchancment over the CCNG1 Library. iCipa was in a position to predict interplay scores, with R2 = 0.43 (Fig. 4E). We attribute the rise in iCipa’s energy to using a coiled-coil background that consists of solely alanine residues on the b-, c– and f– positions. The development in predictive energy appeared in different algorithms to a lesser extent, all of which maintained an R2 < 0.28 (Supplementary Fig. 21).