Cryo-EM constructions of CRF01_AE and CRF07_BC Envs
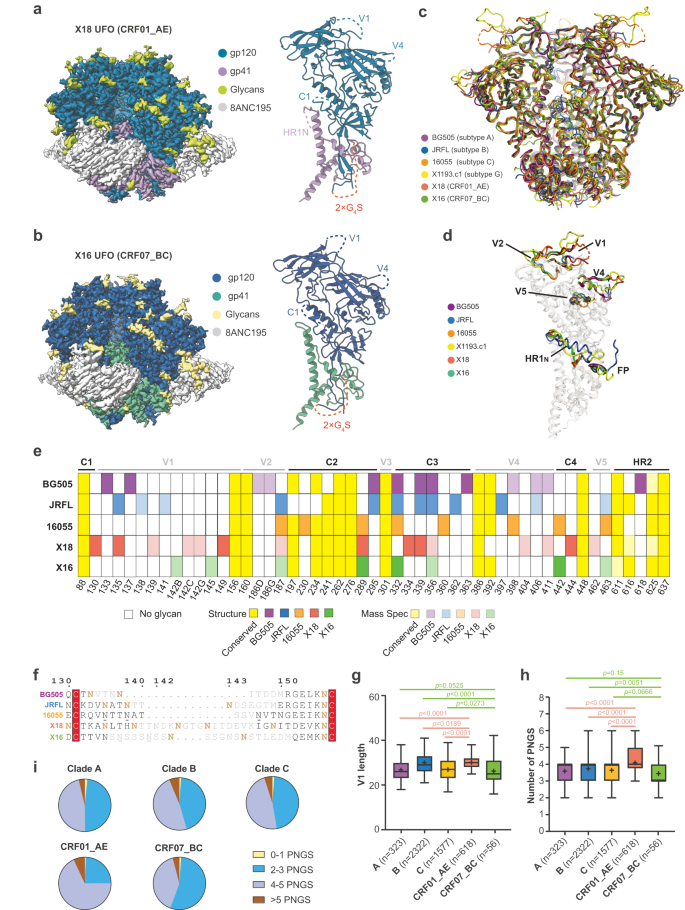
The Env gene sequences representing subtypes CRF01_AE and CRF07_BC have been from virus isolates BJOX018000.e12 (X18, GenBank ID: AIS42911.1) and BJOX016000.e15 (X16, GenBank ID: AIS42666.1), every obtained by means of single genome amplification (SGA) from acute/early HIV-1 contaminated people in China21. To beat Env metastability and create secure, homogeneous Env trimers for structural research, uncleaved prefusion-optimized (UFO) design was utilized to those two Env sequences22, producing X18 UFO (CRF01_AE Env) and X16 UFO (CRF07_BC Env) (Supplementary Fig. 1a). We complexed X18 UFO and X16 UFO with bNAb 8ANC195 to additional stabilize the Env trimers in prefusion state (Supplementary Fig. 1b, c) and decided their cryo-EM constructions at 3.0 Å and three.5 Å, respectively (Fig. 1a, b, Supplementary Fig. 1d–i and Supplementary Desk 1). The general constructions of X18 UFO and X16 UFO undertake closed trimeric conformation in prefusion state, and carefully resemble the Env constructions of consultant strains BG505 (subtype A), JRFL (subtype B), 16055 (subtype C) and X1193.c1 (subtype G) (Fig. 1c)23,24,25,26,27,28. Pair-wise structural alignments revealed low Cα root-mean-square deviation (RMSD) values for each protomers and trimers amongst these Envs (Supplementary Fig. 1j), additional corroborating the general structural homology of Envs throughout subtypes.
a–b Cryo-EM maps (facet views) of X18 UFO (a) and X16 UFO (b) in complicated with 8ANC195 Fab are proven on the left. The reconstituted fashions of X18 UFO (a) and X16 UFO (b) protomers are proven on the suitable, whereby areas invisible in constructions are indicated with dashed traces and labeled. c The atomic fashions of X18 UFO (CRF01_AE) and X16 UFO (CRF07_BC) trimers are proven as ribbon diagrams and superimposed onto the atomic fashions of BG505 SOSIP28 (subtype A, PDB:5CJX), JRFL SOSIP27 (subtype B, PDB: 5FYK), 16055 NFL23 (subtype C, PDB:5UM8) and X1193.c1 SOSIP27 (subtype G, PDB:5FYJ) Env trimers. d Superimposition of X18 UFO, X16 UFO, BG505 SOSIP, JRFL natively cleaved25 (PDB:5FUU), 16055 NFL and X1193.c1 SOSIP Env protomers highlights the conformational variations in surface-exposed variable loops, in addition to FP and HR1N areas. For readability, the superimposed fashions are rendered clear aside from these areas. e PNGS comparability amongst BG505, JRFL, 16055, X18 and X16. PNGS which might be seen in constructions or confirmed by mass spectrometry are represented by completely different shades of indicated colours. These conserved PNGS are coloured yellow. Glycan positions are numbered in response to HXB2 numbering. MS knowledge is both from this research or from earlier publications37. f Sequence alignment is proven for BG505, JRFL, 16055, X18 and X16 V1 loops, with residues invisible in constructions coloured gray. Confirmed PNGS are in daring brown, unconfirmed PNGS are underlined. g–h Field plots of the V1 loop size (g) and PNGS (h) of subtypes A (n = 323), B (n = 2322), C (n = 1577), CRF01_AE (n = 618) and CRF07_BC (n = 56). The field and whisker signify the 25–75% and 5–95% percentile respectively, the median is proven as daring line and the imply is depicted with ‘+’. P values are given by the Mann–Whitney check. i Pie plots depicting the fractions of V1 loops with 0–1, 2–3, 4–5 and >5 PNGS in every HIV-1 subtype. HIV-1 Env sequences are from the Los Alamos HIV sequence database (www.hiv.lanl.gov). Supply knowledge are offered as a Supply Knowledge file.
Regardless of the general structural similarities, native conformational variations exist amongst X18 UFO, X16 UFO and the Envs of consultant strains from different subtypes, most notably within the heptad repeat 1 N-terminal (HR1N, 548-568) area, FP area and surface-exposed hypervariable loops V1, V2, V4 and V5 (Fig. 1d). Whereas the structural variations within the hypervariable loops are per their excessive sequence variations, the conformational variations within the HR1N area is basically attributable to the adoption of native-like trimer designs29, whereby the engineering of proximal areas in SOSIP30, NFL31 or UFO22 design all disrupt the native α-helical conformation of HR1N (Supplementary Fig. 1k, evaluate others to JRFL WT). The FPs are typically ‘uncovered and disordered’ within the closed state of prefusion Env trimers however would rearrange to turn out to be ‘buried’ within the CD4-induced open state of Env32. In step with their closed prefusion conformations, the FPs in X18 UFO and X16 UFO are each ‘uncovered’, akin to the FP conformation in native prefusion Env trimers (Supplementary Fig. 1l, in comparison with JRFL WT). Beforehand, a uncommon ‘buried’ conformation of FP has been captured in a 3.9 Å prefusion construction of a really early transmitted founder virus (CRF01_AE T/F100), which was thought to signify an intermediate between the closed and open states33. Right here, the FP of X18 UFO (additionally from CRF01_AE) didn’t recapitulate the ‘buried’ intermediate state seen in T/F100 SOSIP trimer, presumably due that the UFO trimer just isn’t cleaved between gp120 and gp41 as in SOSIP trimer.
We additionally in contrast the interfaces between 8ANC195 and X18 UFO (CRF01_AE), X16 UFO (CRF07_BC) or BG505 Env (subtype A), and located that amino acid (AA) variations of interface residues among the many three Envs don’t have an effect on the binding of 8ANC195 a lot (Supplementary Fig. 2a–f). Furthermore, we calculated the AA utilization frequencies of CRF01_AE (n = 618) and CRF07_BC (n = 56) viruses in 8ANC195-contacting area and in contrast them to Env signatures related to 8ANC195 sensitivity (Supplementary Fig. 2g), which signifies that the sensitivities of CRF01_AE and CRF07_BC viruses to 8ANC195 wouldn’t considerably differ from different M-group viruses.
Glycosylation options of consultant strains from completely different subtypes
The HIV-1 Envs are closely glycosylated. For neutralization, bNAbs need to accommodate and even particularly acknowledge sure glycans throughout the glycan protect of Env6,34. We thus analyzed the N-linked glycans on X18 UFO and X16 UFO intimately. There are 29~30 potential N-linked glycosylation websites (PNGS) in every protomer of X18 or X16, we noticed 18-20 of them within the cryo-EM constructions (Fig. 1e) and confirmed the existence of different PNGS, most of which find in or close to the structurally invisible variable loops, by means of mass spectrometry evaluation (see strategies). Collectively, the construction and MS evaluation recognized 30 of the 30 PNGS in every protomer of X18 Env and 24 of the 29 PNGS in X16 Env (Fig. 1e). Of those confirmed PNGS, 16 are conserved (current in > 85% of HIV-1 Env sequences, n = 6515) amongst M-group viruses (Fig. 1e, yellow).
As beforehand reported, CRF01_AE and CRF07_BC are recombinants of subtypes A/E and subtypes B/C respectively35,36. We thus in contrast the glycan shields of X18 (CRF01_AE) and X16 (CRF07_BC) to these of consultant strains from subtypes A (BG505), B (JRFL) and C (16055)23,25,37,38 (Fig. 1e). Whereas conserved glycosylation websites typically find within the sequence-conserved C1, C2 and heptad repeat 2 (HR2) areas (Fig. 1e, yellow), a lot of the strain-specific N-linked glycans cluster round hypervariable loops, aside from V3 whereby N301 glycosylation is very conserved (Fig. 1e). Of word, the V1 of X18 (CRF01_AE) is glycosylated at 6 completely different websites, making it probably the most closely glycosylated V1 among the many 5 strains (Fig. 1e, in comparison with 2~3 such websites in different strains). The conserved C3 and C4 areas additionally manifested strain-specific glycosylation patterns, whereby native shifting of glycosylation websites are the commonest manifestations (Fig. 1e). For example, whereas pressure BG505 is glycosylated at N363, this glycosylation web site is shifted to N362 and N360 in pressure JRFL and pressure 16055 respectively (Fig. 1e). In C4 area, N-glycan can also be shifted from N442 in pressure 16055 and X16 to N444 in X18 (Fig. 1e). Notably, native shifting of glycosylation websites could also be a possible technique for HIV-1 Env to evade the popularity of neutralizing antibodies whereas nonetheless sustaining the integrity of glycan protect close by34.
The distinctive options of CRF01_AE V1 area
As famous above, the V1 loop in X18 hosts extra PNGS than different strains (Fig. 1e, f). Furthermore, the V1s of X18 (CRF01_AE) and X16 (CRF07_BC) are considerably longer than these of strains BG505 (subtype A), JRFL (subtype B) and 16055 (subtype C) (Fig. 1f). Subsequent, we sought to discover whether or not these strain-specific options of X18 and X16 V1s could possibly be generalized to corresponding subtypes. We first in contrast the V1 lengths in CRF01_AE (n = 618) and CRF07_BC (n = 56) Envs to these in subtypes A (n = 323), B (n = 2322) and C (n = 1577) Envs (Fig. 1g). The information signifies that subtype CRF01_AE certainly tends to have an extended V1 than subtypes A, B and C (Fig. 1g). Alternatively, the V1 size in CRF07_BC doesn’t deviate a lot from that in subtype A, though it appears to be shorter than that in subtypes B and C (Fig. 1g). We then in contrast the PNGS numbers in V1s of CRF01_AE and CRF07_BC Envs to these of subtype A, B and C Envs and located that the variations between CRF01_AE and subtypes A, B and C are all extremely vital (Fig. 1h). The share of Envs internet hosting 4 or extra V1 glycans reaches 75% in CRF01_AE whereas this quantity solely ranges 50~54% in subtypes A, B and C (Fig. 1i). In the meantime, the typical V1 PNGS quantity in CRF07_BC is decrease than that in subtype B, though the distinction between CRF07_BC and subtype A or C just isn’t statistically vital (Fig. 1h). Persistently, the share of Envs internet hosting 4 or extra V1 glycans in CRF07_BC is decrease (Fig. 1i, 44% in CRF07_BC versus 50~54% in subtypes A, B and C). Thus, the cross-subtype statistical evaluation of M-group HIV-1 Env sequences revealed that the V1 loop of CRF01_AE is considerably longer and glycosylated heavier than the V1s of subtypes A, B and C.
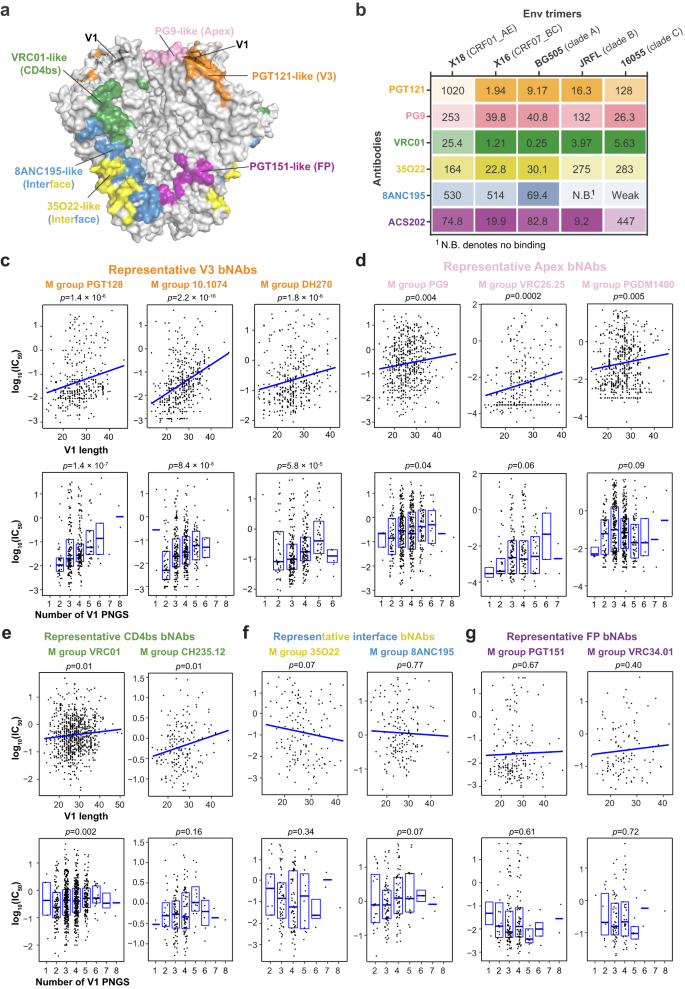
V1 options are related to sensitivities to sure bNAbs
The Env molecular options close to epitope can influence the operate of corresponding bNAbs. For example, prolonged size and better variety of PNGS inside V5 have been correlated with elevated resistance to CD4bs bNAbs, which bind close to the V5 area12. Contemplating that the V1 area locates close to the epitopes of V3, V1/V2 apex and CD4bs bNAbs (Fig. 2a), V1 options like size or glycosylation degree might affect the binding of those bNAbs. To check this speculation, we chosen 1–2 consultant bNAbs from every group of V3, Apex, CD4bs, gp120-gp41interface and FP bNAbs and characterised their interactions with X18 (CRF01_AE), X16 (CRF07_BC) or Envs of subtypes A, B and C consultant strains (Fig. 2b and Supplementary Fig. 3a). We discovered that the affinities between X18 and PGT121 (V3 bNAb), PG9 (Apex bNAb) or VRC01 (CD4bs bNAb) are all clearly decrease than these between different Envs and these bNAbs (Fig. 2b). In distinction, the interactions between X18 and interface bNAbs 8ANC195, 35O22 or FP bNAb ACS202 are all near common as in comparison with different Envs (Fig. 2b). These outcomes counsel {that a} lengthy and closely glycosylated V1 doubtless renders X18 much less delicate to the examined V3, Apex and CD4bs bNAbs.
a Floor illustration of prefusion Env trimer coloured by the indicated bNAbs epitopes, with V1 loops in two neighboring Env protomers proven in cartoon. Notice that the V1 loop locates in proximity to V3, Apex and CD4bs bNAbs epitopes and thus might have an effect on the binding of those bNAbs. b The binding affinities between indicated bNAbs and Env trimers from consultant strains of various subtypes are decided by SPR and proven as OkD (nM). Lighter shades of coloration point out decrease affinity and vice versa. X18 UFO (CRF01_AE) is much less delicate to PGT121 (V3 bNAb), PG9 (Apex bNAb) and VRC01 (CD4bs bNAb) as in comparison with the Envs of consultant strains from different subtypes. c–g Correlations between V1 loop traits and bNAb sensitivities are represented by scatter plots. High: Regression traces have been fitted utilizing the ggplot2 bundle in R (stat_smooth operate). Backside: The field signify the 25–75% percentile and the median is proven as daring line. HIV-1 viruses neutralization knowledge and V1 loop traits of corresponding viruses for consultant V3 bNAbs (c), Apex bNAbs (d), CD4bs bNAbs (e), interface bNAbs (f) and FP bNAbs (g) are ready as described in Strategies and saved in Supply Knowledge. The p values are calculated from Kendall’s tau. The pattern dimension are as following (n represents biologically impartial samples): PGT128 (n = 348), 10.1074 (n = 339), DH270 (n = 366), PG9 (n = 546), VRC26.25 (n = 288), PGDM1400 (n = 570), VRC01 (n = 840), CH235.12 (n = 186), 35O22 (n = 148), 8ANC195 (n = 166), PGT151 (n = 189) and VRC34.01 (n = 110). Supply knowledge are offered as a Supply Knowledge file.
To verify the correlation between the V1 molecular options and the viral sensitivities to V3, Apex and CD4bs bNAbs, we then calculated the affiliation between V1 size (or PNGS quantity) and measured neutralization efficiency of prototypical V3, Apex or CD4bs bNAbs on M-group HIV-1 viruses (Fig. 2c–e). We discovered that the V1 size correlates inversely with the viral sensitives to consultant V3 bNAbs from three completely different lineages (Fig. 2c, prime panels and Supplementary Fig. 3b). Additionally, extra PNGS in V1 strongly correlates with decreased sensitivity to V3 bNAbs PGT128, 10.1074 and DH270 (Fig. 2c, backside panels), although to not PGT121 (Supplementary Fig. 3b).
For prototypical Apex bNAbs PG9, VRC26.25 and PGDM1400, the V1 lengths are additionally inversely correlated with their neutralizing efficiency (Fig. second, prime panels). Some weak correlations between the V1 PNGS numbers and resistance to those bNAbs will also be recognized (Fig. second, backside panels). We additionally discovered that elevated V1 size is linked with decreased sensitivity to VRC01 and CH235.12, two consultant CD4bs bNAbs (Fig. 2e, prime panels), per earlier findings that lengthy V1 and V2 areas can mediate in vivo escape to CD4bs bNAbs39. The correlation between PNGS numbers and CD4bs bNAbs sensitivities is bNAb particular, as a correlation was noticed for VRC01 whereas no correlation was discovered for CH235.12 (Fig. 2e, backside panels). No apparent correlation was recognized between the molecular options of V1 and the neutralizing efficiency of consultant interface or FP bNAbs (Fig. 2f, g), which is conceivable because the epitopes of theses bNAbs all find removed from V1.
The above outcomes thus point out a considerably greater likelihood for CRF01_AE viruses to be proof against V3 and Apex bNAbs, given the distinctive molecular options of their V1s. These outcomes additionally counsel that the regional distributions of subtypes ought to be thought-about in epitopes-focusing vaccine design and the prophylactic utilization of corresponding bNAbs (mentioned later intimately).
bNAb F6 acknowledges a gp120-gp41 interface epitope
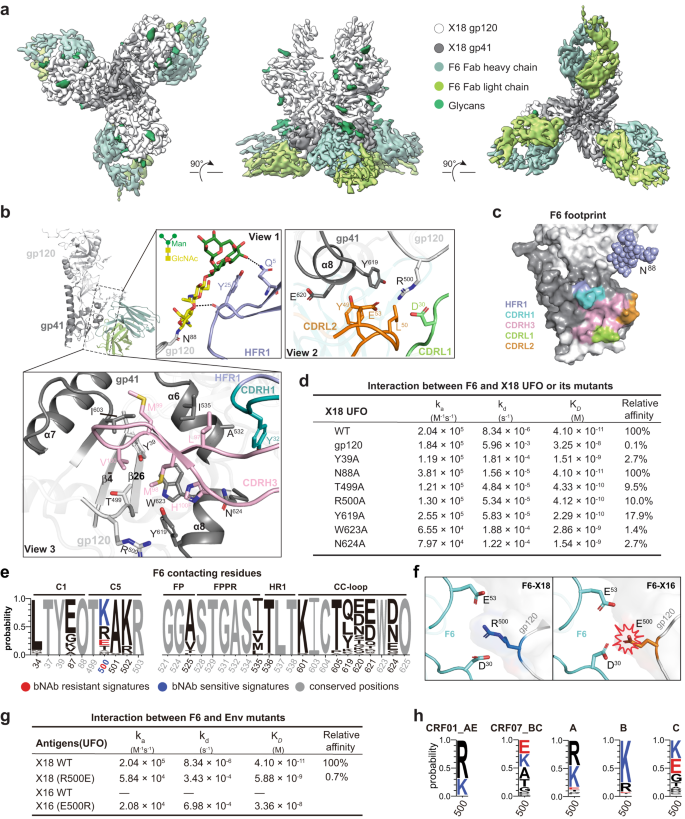
To date, only a few bNAbs are remoted from CRFs-infected donors, limiting our understanding of the host immune response in opposition to CRFs infections. To additional lengthen our antigenic understanding of CRF Envs, we then solved the cryo-EM construction of X18 UFO (CRF01_AE) in complicated with F6, which is the primary bNAb (57% breadth) obtained from a CRF01_AE-infected particular person20. After intensive cryogenic situation optimization, we have been capable of reconstitute a 4.14 Å cryo-EM map of X18 UFO-F6 complicated whereby three F6 bind one X18 UFO trimer (Fig. 3a, Supplementary Fig. 4a–d and Supplementary Desk 1). Within the reconstituted cryo-EM map, the density of the Env apex proximal areas (V1/V2/V3 and a part of C4, V4, V5) is shattered and untraceable, indicating excessive flexibility (Fig. 3a). In distinction, the densities of different Env areas, in addition to the variable domains of F6, are all sufficiently good for unambiguous mannequin constructing, particularly on the interface after native refinement (Supplementary Fig. 4e–g). To facilitate the mannequin constructing course of, we additionally solved a 3.3 Å crystal construction of bNAb F6 alone (Supplementary Desk 2). This construction and the X18 UFO construction solved on this research have been then docked into the EM map and additional refined to generate the atomic mannequin of the X18 UFO-F6 complicated.
a Cryo-EM reconstitution of X18 UFO trimer in complicated with bNAb F6. Proven are the highest (left), facet (center) and backside (proper) views. b The interfaces between X18 UFO protomer and F6 Fab are depicted with an total sideview (left) and detailed in three close-up views (boxed). The gp120 and gp41 subunits of X18 UFO are proven as gentle and darkish gray ribbons, and the HFR1, CDRL1, CDRL2, CDRH1 and CDRH3 of F6 Fab are proven as purple, inexperienced, orange, cyan and pink ribbons in close-up views. Glycan N88 and the important thing residues concerned in interactions are proven as stick fashions. The Env residues are labeled in response to HXB2 numbering and CDR residues are labeled following Kabat numbering. c Footprint of bNAb F6 on the floor presentation of HIV-1 Env protomer (gp120: white, gp41: gray). Env surfaces that work together with HFR1, CDRH1, CDRH3, CDRL1 and CDRL2 of F6 are coloured purple, cyan, pink, inexperienced and orange respectively. d Interplay kinetics between F6 and X18 UFO trimer (or its indicated mutants). e Amino acid (AA) signatures in bNAb F6 contacts are recognized by Fisher’s check from the dataset saved in Supply Knowledge and displayed in WebLogo plots whereby letter peak represents AA frequencies. AAs related to sensitivity and resistance to F6 are coloured blue and crimson, respectively. “O” denotes an Asn (N) in a PNGS motif. f Left: R500 from the gp120 of X18 types salt bridges with D30 and E53 from the sunshine chain of F6. Proper: Change of R500 to E500 would result in electrostatic repulsion amongst E500, D30 and E53, thus explaining the failure of X16 to bind F6. g Binding of F6 to wild-type X18 UFO is sort of abrogated by single R500E mutation, and single E500R mutation renders X16 UFO aware of F6. h WebLogo plots of the AA frequencies at residue 500 in subtypes A (n = 323), B (n = 2322), C (n = 1577), CRF01_AE (n = 618) and CRF07_BC (n = 56) Envs. Env sequences are from the Los Alamos HIV sequence database (www.hiv.lanl.gov). Supply knowledge are offered as a Supply Knowledge file.
Akin to different bNAbs, the epitope of F6 additionally includes N-glycans. Particularly, the conserved N88 glycan packs in opposition to the heavy chain framework area 1 (HFR1) of F6 and contributes ~350 Å2 buried epitope areas (Fig. 3b, close-up view 1). When N88 is mutated to Ala, the affiliation fee between F6 and X18 UFO roughly doubles, whereas the disassociation fee additionally accelerates about twofold, leaving the OkD largely unchanged (Fig. 3d and Supplementary Fig. 5a). Such phenomenon is paying homage to 3BC315, a gp41-binding bNAb remoted from clade B contaminated people, which additionally exhibits synchronously elevated on- and off-rates within the absence of glycan40. Therefore, as beforehand proposed, though N88 glycan just isn’t required for F6 and 3BC315 to bind and will even sterically prohibit their entry, it could work as a ‘clasp’ to assist maintain antibodies in place as soon as they’re sure40.
F6 additionally makes use of its complementarity-determining area (CDR) 1 and a pair of in gentle chain (CDRL1 and CDRL2) to concurrently have interaction the α8 helix of gp41 and the C-terminus of gp120 (Fig. 3b, close-up view 2). Of all of the Env residues at this interface, R500 from gp120 performs a central function. The aliphatic a part of R500 and the fragrant ring of Y619 from α8 helix type a hydrophobic cluster with the sidechains of Y49 and L50 from CDRL2, zipping α8 (gp41), the C-terminus of gp120 and CDRL2 collectively. In the meantime, R500 additionally types electrostatic interactions with E53 from CDRL2 and D30 from CDRL1, additional strengthening the adhesion of F6. When R500 was mutated into Ala, a 90% affinity lower was noticed, corroborating its significance (Fig. 3d and Supplementary Fig. 5a). In comparison with the interface between the Env and the sunshine chain CDRs, the interface between the Env and the heavy chain CDRs (CDRHs) is even bigger, and the interplay is especially pushed by hydrophobic packings (Fig. 3b, close-up view 3). The CDRH3 adopts an anti-parallel β-sheet conformation and side-wedges right into a hydrophobic cleft shaped by residues I535, I603, Y619 and W623 from α6-α8 helices of gp41 and residues Y39 and T499 from ({{{{{rm{beta }}}}}}bar{4}/{{{{{rm{beta }}}}}}26) sheets of gp120 (Fig. 3b, close-up view 3). Moreover CDRH3, Y32 from CDRH1 additionally types hydrophobic interactions with the facet chain of A532 within the α6 helix (gp41) (Fig. 3b, close-up view 3). These structural analyses clearly present that F6 concurrently have interaction residues and glycans from each gp120 and gp41, thereby categorizing it into the gp120-gp41 interface bNAbs (Fig. 3c). Certainly, Ala mutations on gp120 residues like Y39, T499 and R500 typically result in 90% to 97% affinity lower, and Ala mutations on gp41 residues like Y619, W623 and N624 additionally lead to 82% to 99% affinity lower, additional validating the gp120-gp41 spanning epitope of F6 (Fig. 3d and Supplementary Fig. 5a).
Env signatures related to viral sensitivity to bNAb F6
To offer additional info for immunofocusing vaccine design and potential scientific utilization of F6, we then tried to retrieve the Env signatures related to F6 sensitivity. Utilizing the out there F6 neutralization dataset20, we calculated the correlations between viral molecular signatures close to F6 epitope (e.g., AAs utilization and glycan signatures) and the sensitivities of corresponding viruses to F6. We discovered that Ok at residue 500 in gp120 is related to F6 sensitivity whereas E at this place is linked to F6 resistance (Fig. 3e). On condition that Ok and R are chemically comparable, we postulated that R at this place shall even be related to F6 sensitivity though the present correlation just isn’t statistically vital, presumably as a result of small dimension of current neutralization dataset. Certainly, wildtype X18, which bears an R at residue 500, binds F6 with excessive affinity (Fig. 3g, OkD at 41 pM). In distinction, wild kind X16, which bears an E at residue 500, doesn’t bind F6 in any respect (Fig. 3g and Supplementary Fig. 5a). As proven above, R500 on Env types salt bridges with D30 and E53 from F6 (Fig. 3b). Therefore, the change of R500 to E500 would result in electrostatic repulsion amongst E500, D30 and E53 (Fig. 3f), thereby explaining the failure of X16 to bind F6 (Fig. 3g). To additional assist the revealed F6 sensitivity signatures, a R500E mutation nearly eliminates the binding of F6 to X18 (0.7% of unique affinity), whereas a E500R mutation endows X16 responsiveness to F6 (Fig. 3g and Supplementary Fig. 5a).
Subsequent, we calculated the AA frequencies at residue 500 in subtype A, B, C, CRF01_AE and CRF07_BC Envs to foretell the sensitivities of various HIV-1 subtypes to F6. The upper frequency of F6 resistant signature at residue 500 (i.e., E500) in CRF07_BC and subtype C viruses means that viruses from these two subtypes usually tend to be F6 resistant as in comparison with viruses from CRF01_AE, subtypes A and B (Fig. 3h).
Comparability of F6 to different gp120-gp41 interface bNAbs
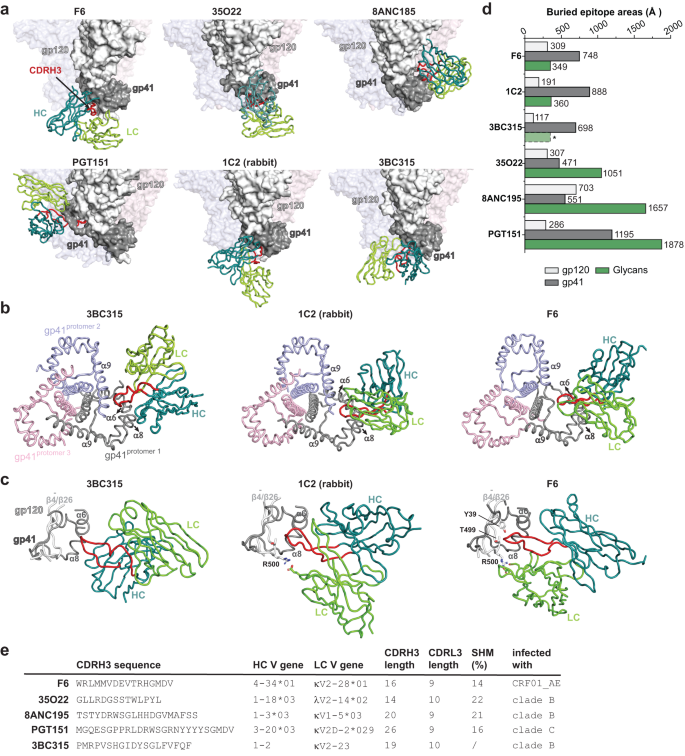
Subsequent, we in contrast F6 to 35O22, 8ANC195, PG151 and 3BC315, bNAbs which were proven to bind close to the interface between gp120 and gp4125,28,38,40. The epitope of F6 is completely different from that outlined by 8ANC195 and 35O22, two prototypical gp120-gp41 interface bNAbs, and can also be distinct from the epitope of PGT151, a consultant FP bNAb (Fig. 4a). In the meantime, the binding place of F6 on Env is much like that of 3BC315, though their approaching angles and orientations of heavy and light-weight chains are completely different (Fig. 4a). We additionally in contrast F6 to 1C2, a cross-reacting antibody (87% breadth) elicited by heterologous Env trimer-liposome prime:boosting in rabbits41 (Fig. 4a). The epitope of 1C2 overlaps that of F6 by round 953 Å2 (Supplementary Fig. 5b) and their approaching angles are very comparable (Fig. 4a). Nonetheless, though F6, 3BC315 and 1C2 acknowledge overlapping areas on Env floor, variations are apparent amongst them. Within the context of Env trimer, 3BC315 and 1C2 work together with gp41 from two neighboring protomers concurrently, whereas F6 solely work together with gp41 from one protomer (Fig. 4b). Furthermore, F6 makes intensive and key interactions with gp120 (Fig. 3b–g), whereas 1C2 solely make restricted interactions with gp120 and 3BC315 barely interacts with gp120 residues40 (Fig. 4c). The completely different participation of gp120 additionally explains the variations of their neutralization mechanism (see dialogue later).
a The binding mode of F6 is in comparison with these of different interface bNAbs. The constructions of F6-Env and 8ANC195-Env complexes are from this research, the constructions of 35O22-Env, PGT151-Env and 1C2-Env complexes are based mostly on PDB: 4TVP, 5FUU and 6P6525, 38, 41. The constructions of 3BC315-Env are obtained by becoming the 3BC315 crystal construction (PDB: 5CCK) and the BG505 Env mannequin (PDB: 4TVP) into the 9 Å cryo-EM map of 3BC315 in complicated with BG505 SOSIP (EMD-3067)40. In every panel, solely the variable areas of 1 bNAb are proven, the gp120 and gp41 of the key interacting Env protomer are proven as white and gray floor shows whereas the opposite two protomers are rendered clear for readability. b Comparability of the interactions between gp41 and 3BC315, 1C2 and F6. 3BC315 and 1C2 work together with the α6 and α8 from one protomer (gray), in addition to α9 from the neighboring protomer (purple). F6 solely work together with the α6 and α8 from the identical protomer. c Comparability of the interactions between gp120 and 3BC315, 1C2 and F6. 3BC315 barely interacts with gp120 residues. The sunshine chain (LC) of 1C2 makes restricted interplay with R500 from gp120. The sunshine chain (LC, CDRL1 and CDRL2) and heavy chain (HC, CDRH3) of F6 make intensive and essential interactions with R500, T499 and Y39 from gp120. Notice that the CDRH3 of F6 inserts a lot deeper to achieve ({{{{{rm{beta }}}}}}bar{4}/{{{{{rm{beta }}}}}}26) (gp120) than that of 3BC315 or 1C2. d Bar graphs denoting gp120, gp41 and glycan interacting floor areas on Env for every indicated bNAb. *The contribution of glycans to 3BC315 epitope is inferred slightly than calculated from the docking mannequin. e The CDRH3 sequences, heavy and light-weight chain V gene utilization, CDRH3 and CDRL3 lengths, somatic hypermutation (SHM) charges and the origins of the indicated gp120-gp41 interface bNAbs. Supply knowledge are offered as a Supply Knowledge file.
The contributions of N-linked glycans to epitopes are additionally completely different amongst these antibodies (Fig. 4d). Within the context of Env protomer, F6 buries 309 Å2 and 748 Å2 epitope areas on gp120 and gp41 respectively, whereas N-linked glycans solely contribute 349 Å2 to its buried epitope space and seem like dispensable (Figs. 3d, 4d). In distinction, glycans are indispensable for the binding of 35O22, 8ANC195 and PGT15142,43,44 and the contributions of glycans (1051 Å2, 1657 Å2 and 1878 Å2) to 35O22, 8ANC195 and PGT151 epitopes all outweigh the contributions of protein half (748 Å2, 1254 Å2 and 1481 Å2) (Fig. 4d). Akin to F6, the epitopes of 1C2 and 3BC315 solely obtained restricted contributions from glycans (Fig. 4d). Of word, the epitope of F6 locates close to one of many two main practical gaps within the steady glycan protect on Env floor. As the world close to F6 epitope just isn’t obscured by N-glycan shielding, a viable host recognition of this space is assumed. Certainly, the truth that this space is acknowledged by F6 and 3BC315, two bNAbs from completely different clonal linages and distinct HIV-1 an infection background (Fig. 4e), demonstrates that the hosts can successfully respect this conserved vulnerability regardless of their various genetic background. In the meantime, the truth that this space can also be effectively captured by vaccination induced immunity in rabbits (1C2) additional means that this space could also be a possible goal web site for immunofocusing vaccine design.
F6 weakens protomers engagement and triggers trimer disassembly
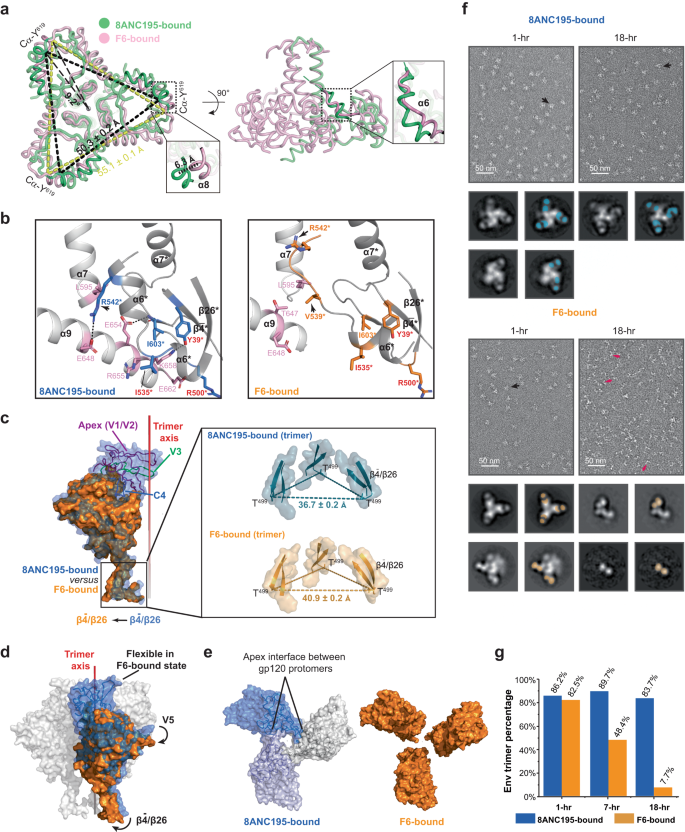
As talked about above, the Env apex proximal areas grew to become extremely versatile upon F6 binding (Fig. 3a). To learn the way F6 triggered this, we in contrast the construction of X18 UFO in F6-bound state to that in 8ANC195-bound state. In gp41, we noticed a 6.3 Å motion of α8 helix and 5.6 Å motion of α6 helix, plus a de-spiral on the C-terminus of α6 in F6-bound state as in comparison with 8ANC195-bound state (Fig. 5a, close-up views). In 8ANC195-bound state, intensive inter-protomer interactions are noticed not solely among the many central helices (α7), but in addition between the α9 helix and the α6 helix from the neighbor protomer (Fig. 5b, left). In F6-bound state, though the interactions amongst α7 helices stay largely unchanged (Supplementary Fig. 6a), the motion of α6 in the direction of F6 and its de-spiral break the hydrophobic (e.g., L595 and aliphatic a part of R542*, * denotes components from neighbor protomer) and hydrophilic (e.g., E648 and R542*) inter-protomer packings between α7/α9 and α6* helices (Fig. 5b, evaluate proper to left). In the meantime, the involvement of Env residues Y39*, R500* and I535* in F6 binding disrupts their unique interplay with neighbouring α9 helix residues K658, E662 and R655 (Fig. 5b, evaluate proper to left), which, along with the steric clashes rendered by F6 (Supplementary Fig. 6b), destabilize the C-terminus of the α9 helix and result in additional inter-protomer packing loss between α9 and α6* helices (Fig. 5b). To summarize, the F6-induced positional and conformational modifications in α6 and α9 helices destabilize the interactions among the many Env protomers on the base finish (Fig. 5b). Consequently, the F6-bound gp41 trimer shows a 9.2o clockwise rotation and ~5 Å dilation on the base finish, as in comparison with 8ANC195-bound state (Fig. 5a).
a Left: backside view of aligned gp41 trimers, gp41 trimer shows a 9.2o clockwise rotation and a ~5 Å dilation in F6-bound state (pink) as in comparison with 8ANC195-bound state (inexperienced). Proper: facet view. The 2 boxed close-up views depict the ~6 Å translocation of α8 helix and the de-spiral of α6 helix in F6-bound state as in comparison with 8ANC195-bound state. b F6 Binding destabilizes the interactions amongst Env protomers on the base finish of gp41. Residues concerned in inter-protomer packings in 8ANC195-bound Env trimer are proven as pink (protomer 1) and blue (protomer 2) sticks (left). The identical residues are proven as pink and orange sticks in F6-bound state (proper). F6-binding Env residues are labeled crimson. Arrows point out residues displaying massive positional modifications in F6-bound state. Structural components and residues of protomer 2 are denoted with * and electrostatic interactions are indicated with black dashes. c Aligned gp120 protomers spotlight the outward motion of ({{{{{rm{beta }}}}}}bar{4}/{{{{{rm{beta }}}}}}26) (indicated with an arrow) and the lacking of apex proximal areas (V1/V2/V3 and a part of C4, V4, V5) in F6-bound state (orange) as in comparison with 8ANC195-bound state (blue). Field: close-up views of the ({{{{{rm{beta }}}}}}bar{4}/{{{{{rm{beta }}}}}}26) sheets in trimer context. d Sideview of aligned gp120 trimers in floor representations suggests the potential transmission route of conformational modifications. F6-binding induced rotational actions in ({{{{{rm{beta }}}}}}bar{4}/{{{{{rm{beta }}}}}}26) and V5 areas are indicated with black arrows. e Left: the V1/V2 area from completely different gp120 protomers interacts with one another in 8ANC195-bound state. Proper: the identical interface is lacking in F6-bound state as a result of F6-binding induced destabilization of apex proximal areas. f nsEM micrographs of 8ANC195-bound and F6-bound X18 UFO at 1-h and 18-h. Consultant Fab-bound Env trimers and Fab-bound gp120-gp41 protomers are pointed with black and crimson arrows respectively. Proven beneath every nsEM micrograph are the consultant 2D class averages of corresponding samples and the places of Fabs in every class are highlighted with blue (8ANC195) or orange (F6) blocks on the suitable. g Share of intact Env trimers below nsEM on the indicated time factors. Knowledge for this plot are proven in Supplementary Desk 3.
In gp120, a big outward motion (relative to the trimer axis) was noticed within the ({{{{{rm{beta }}}}}}bar{4}/{{{{{rm{beta }}}}}}26) sheets (Fig. 5c, area boxed in black). Consequently, whereas the 8ANC195-bound X18 UFO measures ~37 Å between the Cα of T499 on adjoining protomer, the identical distance measures ~41 Å in F6-bound state (Fig. 5c, close-up view in trimer context). Notably, the gp120 protomer shows clockwise rotation from the sideview upon F6 binding (Fig. 5d, rotational motion indicated with arrows), which doubtless transmits the disengagement of protomers close to the bottom (Fig. 5b, c) all the way in which to the apex and ultimately results in the destabilization of the apex proximal areas (V1/V2/V3 and a part of C4, V4, V5). Notably, the V1/V2/V3 areas are the place the gp120 protomers have interaction with one another, and their destabilization would loosen the interplay amongst gp120 protomers on the apex (Fig. 5e).
Therefore, the binding of F6 weakens the inter-protomer packings at each the bottom and the apex of Env trimer (Fig. 5a–e), resulting in the potential for inducing Env trimer disassembly. To check for such chance, we then carry out comparative adverse stain EM (nsEM) evaluation of 8ANC195-bound or F6-bound X18 UFO trimers at completely different time factors. Within the presence of 8ANC195, ~90% of the particles stay to be intact 8ANC195-bound Env trimers over an 18-h interval (Fig. 5f, g and Supplementary Desk 3). In distinction, whereas ~83% F6-bound X18 UFO seems to be intact trimers at the start (1-h), the share of intact trimers decreases to ~48% by the top of seven h, and to lower than 10% by the top of 18 h (Fig. 5f, g and Supplementary Desk 3). In the meantime, 2D class averages present that a lot of the particles have disassembled into F6-bound gp120-gp41 protomers by the top of 18 h (Fig. 5f). Notably, F6-induced disassembly was additionally noticed for Q769 Env (subtype A) (Supplementary Fig. 6c). These outcomes clearly display that F6 binding would first destabilize the apex proximal areas and ultimately result in Env trimer disassembly, and our cryo-EM construction of F6-bound X18 UFO trimers has captured an intermediate state alongside the trail of such induced Env disassembly (Fig. 3a).
Neutralization mechanism of F6
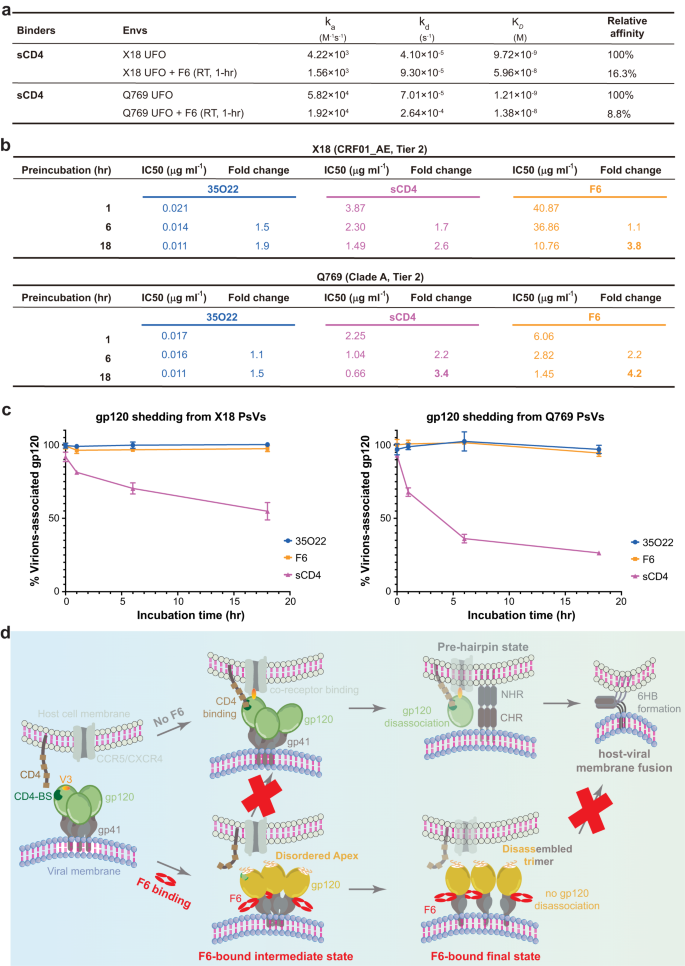
Subsequent, we sought to disclose the neutralization mechanism of F6. In step with the destabilization of trimer apex proximal areas, the Apex bNAbs PGT145 and PG9 acknowledge the F6-bound Env trimer with markedly lowered affinities than to the unbound Env (Supplementary Fig. 7a, b). Such remark not solely confirms that the binding of F6 is actually destabilizing the trimer apex, but in addition signifies that destabilization of corresponding areas would hinder the binding of proteins that acknowledge these areas. Notably, the V1/V2 stem, β20/β21 hairpin and the V5 loop all play essential roles in receptor CD4 engagement32,45 (Supplementary Fig. 7c–e). Certainly, a single mutation at W427 in β20/β21 hairpin is ample to abrogate CD4 binding and render HIV-1 virus non-infectious46. Upon F6 binding, the V1/V2 areas (117–208 aa), the β20/β21 hairpin (421–439 aa) and a part of the V5 loop (459–463 aa) all grew to become destabilized (Supplementary Fig. 7f). Therefore, it’s conceivable that F6-induced destabilization of those areas would inevitably harm their interplay with CD4 (Supplementary Fig. 7d). Persistently, drastic affinity lower was noticed between soluble CD4 (sCD4) and Env when F6 is current (Fig. 6a and Supplementary Fig. 7b). Therefore, F6-induced destabilization of apex proximal areas would hinder the binding of receptor CD4, thereby representing one potential neutralization mechanisms of F6.
a Interplay kinetics between sCD4 and Envs or F6-bound Envs. For the measurement of the latter, X18 UFO or Q769 UFO was solely incubated with F6 for 1 h to preclude the interference of F6-induced trimer disassembly (F6-induced trimer disassembly just isn’t outstanding at 1-h time level). b Pseudotyped X18 or 769 virus was incubated with 35O22 IgG, sCD4 or F6 IgG for indicated durations, earlier than being added to TZM-bl cells. Fold change (lower) in IC50 is calculated by normalizing in opposition to the IC50 of normal 1-h incubation. IC50 values of F6 lower considerably throughout the extended incubation, indicating that Envs are disabled by F6 over time, which is per F6-induced trimer disassembly. c Pseudotyped X18 or 769 virions have been incubated alone or with 35O22, sCD4 or F6 for indicated durations. Incubated virions have been pelleted and gp120 remaining related to virions was detected by ELISA. The quantity of gp120 remaining related to antibody handled virions are normalized in opposition to the untreated pattern (100%). sCD4 and 35O22 function constructive and adverse management respectively. Knowledge are proven as means ± SD (n = 3 biologically impartial experiments). Underlying knowledge for this panel are given in Supplementary Desk 4. d Schematic diagram illustrating the twin neutralization mechanism of F6: (1) F6 binding first destabilizes the apex proximal areas of Env (F6-bound intermediate state), thereby hindering the binding of CD4 receptor; (2) F6-induced destabilization of trimer apex and gp120-gp41 protomers disengagement on the trimer base lastly result in the disassembly of Env trimer into gp120-gp41 protomers (F6-bound remaining state), disassembled Env trimer loses its potential to type fusogenic six-helical bundle (6HB) and thus can’t mediate environment friendly host-viral membrane fusion.
Destabilization of apex proximal area is simply the preliminary occasion triggered by F6. As time goes by, the disengagement of gp120-gp41 protomers at each the trimer apex and base would ultimately result in the disassembly of Env trimers into gp120-gp41 protomers (Fig. 5), thereby hindering host-viral membrane fusion that depends completely on trimeric Env3. Notably, 3BC315 and 1C2, the 2 antibodies that acknowledge comparable areas as F6 on Env floor, additionally induce trimer disassembly and such irreversible disassociation of Env trimers into gp120-gp41 protomers has been correlated with their higher obvious neutralization efficiency at longer incubation time40,41. We thus additionally monitored the neutralization efficiency of F6 over an 18-h interval utilizing an analogous pre-incubation neutralization assay40. For each X18 and Q769 viruses, the obvious IC50 of F6 improved 3.8- and 4.2-fold respectively resulting from F6-induced trimer disassembly over the 18-h incubation, even higher than that of sCD4 (2.6- and three.4-fold, IC50 decreases resulting from sCD4-induced gp120 shedding) (Fg.6b and Supplementary Fig. 8). On condition that 3BC315 has been discovered to additionally speed up gp120 shedding, we checked if F6 might induce the shedding of gp120 from X18 and Q769 viral surfaces as effectively. No apparent gp120 shedding was noticed within the presence of F6 or 35O22, whereas sCD4 effectively induces gp120 shedding as a constructive management (Fig. 6c and Supplementary Desk 4). Therefore, F6 binding would result in the irreversible decay of practical Env trimers into gp120-gp41 protomers on viral surfaces, thereby representing one other potential neutralization mechanisms of it.