Metagenomics requires stringent experimental processes and bioinformatic filtering standards to precisely detect pathogens
The dimensions and complexity of metagenomic sequencing knowledge, in addition to the chance of contamination or pathogen misassignment, necessitate strict experimental and computational protocols to make sure that detected microbes are actually current. We developed procedures that vastly cut back the prospect of calling false positives by (i) utilizing each unfavorable and optimistic controls, (ii) figuring out intersample contamination, and (iii) creating stringent bioinformatic procedures that prioritize specificity over sensitivity (Fig. 1). As a result of our protocols developed over the course of the research, we define our suggestions and the proportion of the 593 complete samples sequenced through metagenomics to which every process was utilized (Supplementary Desk 1).
Experimentally, we developed procedures to each mitigate the chance of and establish potential circumstances of contamination occurring within the laboratory. First, we extracted plasma samples in batches alongside non-template controls (i.e., water controls) for 574 (96.8%) samples. We designed batches to attenuate the circumstances the place samples recognized to be optimistic for a specific pathogen, reminiscent of Lassa virus (LASV), had been extracted or sequenced with samples recognized to lack the pathogen. Earlier than synthesizing cDNA or making ready sequencing libraries, we added a unfavorable management (i.e., RNA remoted from K562 lymphoblast cells) and a optimistic management (i.e., RNA from viral seed inventory spiked into RNA remoted from K562 lymphoblast cells or RNA from a beforehand sequenced plasma pattern recognized to include a selected virus) for 585 (98.7%) and 509 (85.8%) samples, respectively. At this stage, we additionally added sample-specific RNA spike-ins utilizing the Exterior RNA Controls Consortium (ERCC) sequences for every of 508 (85.7%) samples, together with all samples in batches of 12 or extra, growing the likelihood of detecting any downstream cross contamination21. We sequenced the vast majority of samples with combinatorial twin indexes (CDIs), though we used distinctive twin indexes (UDIs) for the one batch sequenced on the NovaSeq 6000 system (99 or 16.7% of samples) to attenuate the chance of misclassification as a result of index hopping.
Computationally, we selected common, strict filtering standards to investigate the ensuing knowledge. We first discarded samples that displayed proof of potential cross-contamination through the ERCC spike-ins (7 of 560 samples; Supplementary Fig. 1A). We then ensured that the anticipated viral genomic materials was recognized within the optimistic controls through the metagenomic classification instrument Microsoft Premonition22 (Supplementary Desk 2). Subsequent, to name a virus current in a pattern, we required it to have (i) at the very least 5 reads assigned to it by Microsoft Premonition; (ii) a larger % of reads assigned to it than assigned to the identical species in any (a) extraction-batch-specific non-template controls, (b) sequence-batch-specific optimistic controls, excluding the spiked in viral genomic materials, and (c) sequence-batch-specific unfavorable controls; and (iii) genome meeting of Microsoft Premonition hits with a threshold of at the very least 10% of the reference genome measurement (Supplementary Information 1, Supplementary Fig. 2). Thus, we mixed a extremely delicate, however much less particular, probabilistic classification instrument with a extremely particular, however much less delicate contig meeting step to assign pathogens to samples.
We assessed the sensitivity and specificity of our metagenomic pipeline relative to scientific RT-qPCR testing standing by utilizing knowledge from the cohort of people suspected of LF. A optimistic Lassa virus (LASV) scientific take a look at was outlined because the amplification of both the GPC gene or the L gene through the commercially out there Altona assay23,24. Prior scientific RT-qPCR standing is an imperfect floor fact, as (i) genome degradation can happen between scientific testing and subsequent sequencing and (ii) RT-qPCR can yield false unfavorable outcomes for samples containing extremely numerous viruses, reminiscent of LASV. Furthermore, we count on PCR to be extra delicate than metagenomics as a result of target-specific amplification25,26. However, we discovered that the Premonition-based thresholds yielded a sensitivity of 91.7% and a specificity of 91.6%; the extra requirement of contig meeting diminished sensitivity to 35.4% however elevated specificity to minimally 96.8% (Supplementary Fig. 1B). The imperfect specificity was attributable to three samples that had been RT-qPCR-negative however optimistic through sequencing. Two of those samples yielded full, equivalent LASV genomes (98% and 99% full), whereas the third pattern yielded a partial genome. We extensively queried these samples and re-tested them through RT-qPCR (Supplementary Word, Supplementary Fig. 3), finally concluding that they had been most certainly diagnostic false negatives, a recognized problem in LASV molecular detection27,28. In abstract, our metagenomic protocols demonstrated excessive specificity for figuring out pathogens in a given pattern.
Metagenomics identifies Lassa virus co-infections of prognostic significance in addition to viral etiologies of Lassa-like sickness
We first used our metagenomic strategy on 560 samples collected from population-level surveillance of people with signs per LF, a viral hemorrhagic fever brought on by LASV that’s endemic to West African nations. We analyzed 458 RT-qPCR-positive and 95 RT-qPCR-negative samples to establish viral co-infections of prognostic significance, uncover viral etiologies of LF-like scientific syndromes in Nigeria, and characterize LASV range. The samples had been collected between 2017 and 2020, span sufferers seen in 15 of 36 states and the Federal Capital Territory, and embody 220 samples from which we beforehand reported LASV genomes14 (Desk 1).
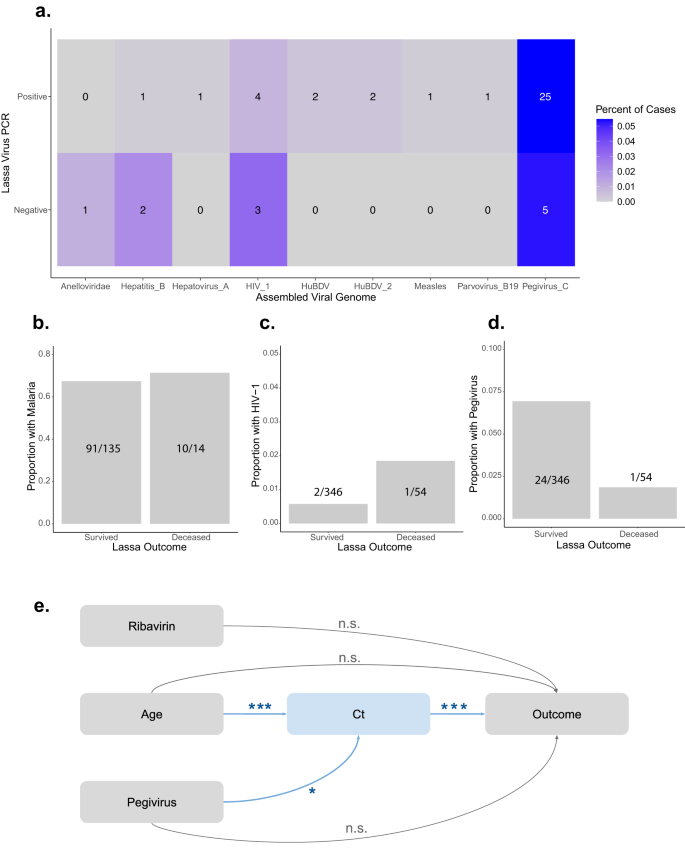
We analyzed the metagenomics reads for different viral pathogens current in our LASV-positive samples, utilizing the filters described above to prioritize specificity over sensitivity. We discovered that 7.8% (36/458) of LASV sufferers had a viral co-infection with at the very least one of many following viruses: hepatitis B, hepatovirus A, human blood-associated dicistrovirus (HuBDV), human immunodeficiency virus 1 (HIV-1), measles, parvovirus B-19, pegivirus C, and an unclassified dicistrovirus that we suggest to call human blood-associated dicistrovirus 2 (HuBDV-2) (Fig. 2a). One pattern was multiply co-infected with each hepatitis B and pegivirus C (Supplementary Information 1). We moreover recognized viruses in 13.7% (13/95) of the RT-qPCR-negative samples, together with LASV as beforehand mentioned, in addition to anellovirus, hepatitis B, HIV-1, and pegivirus C (Fig. 2a). One LASV-negative pattern was multiply co-infected, with anellovirus, LASV (i.e., this pattern was the PCR false unfavorable that produced a partial genome), and pegivirus C.
a Metagenomics identifies Lassa virus (LASV) and non-LASV pathogens in 553 people presenting with signs of Lassa Fever (LF). % (shade scale) and quantity (reported in field) of RT-qPCR-positive (458 samples) or RT-qPCR-negative (95 samples) circumstances containing the next non-LASV pathogens, which had been every present in at the very least one pattern: anelloviridae, hepatitis B, hepatovirus A, human immunodeficiency virus 1 (HIV_1), human blood-associated dicistrovirus (HuBDV), HuBDV-2, measles, parvovirus B19, and pegivirus C. b–d The proportion of surviving or deceased LASV-positive people who had been co-infected with malaria (B), HIV-1 (c), or pegivirus C (d). e Causal directed acyclic graph of hypothesized relationships between ribavirin remedy, age, pegivirus C co-infection standing, LASV cycle threshold (Ct) worth, and outcomes. Arrows are annotated with adjusted p-values produced through multivariate linear (age + pegivirus → Ct; p = 0.0007 for age and p = 0.023 for pegivirus) and logistic (age + Ct + pegivirus + ribavirin → end result; p = 1.85 × 10−12 for Ct) regression fashions. ***p < 0.001. *p < 0.05. n.s. not vital.
As a result of co-infections had been frequent amongst LASV-positive samples, we investigated whether or not they performed a job in LASV outcomes. We analyzed probably the most frequent co-infections (i.e., pegivirus C, HIV-1, and clinically identified malaria) alongside demographic data (i.e., age, intercourse, and being pregnant standing), scientific covariates (i.e., diagnostic Ct and ribavirin remedy standing), and outcomes (i.e., survived or deceased) for 400 LASV-positive people (Desk 2). We performed univariate logistic regression and located that diagnostic Ct worth (p < 0.001) and receipt of ribavirin (p = 0.01) had been considerably related to outcomes, whereas age (p = 0.06) and co-infection with pegivirus C (p = 0.18) trended in direction of an affiliation (Desk 2, Fig. 2b–d, Supplementary Fig. 4A–E). In the meantime, malaria co-infections, which had been recognized in 101 people, weren’t related to outcomes (p = 0.76).
We performed multivariate analyses with the 4 variables that had been related to LASV outcomes at p < 0.25. Prior literature means that these variables work together with outcomes and with each other in complicated methods29,30,31,32,33. For instance, Ct is a measure of the interaction between the host immune system and the virus, which can be affected by age34 or co-infections, however Ct can’t be affected by ribavirin remedy since Ct is measured on the time of analysis earlier than remedy is begun. We developed a causal directed acyclic graph35 (DAG; Fig. 2e), knowledgeable by our univariate analyses and former work29,30,31,32,33, and performed multivariable linear and logistic regression. Age and pegivirus co-infection had been vital predictors of Ct (Fig. 2e, Desk 3, Supplementary Fig. 4G); nonetheless, they weren’t related to the result when controlling for Ct (Fig. 2e, Desk 3, Supplementary Fig. 4F). We subsequently concluded that the impact of age and of pegivirus co-infection standing on the result is mediated by Ct36. We decided that the common causal mediation results of age (p = 2 × 10−16) and of pegivirus co-infection standing (p = 0.02) on end result had been vital through bootstrapping (Supplementary Desk 3, Supplementary Fig. 4H, I). Importantly, we confirmed that there was no relationship between pegivirus C and LASV detection, i.e., as a result of competitors for sequencing reads (Fig. 2a; Supplementary Fig. 4J). Although we can not exclude the opportunity of unknown or unmeasured confounding variables, we computed the mediational E-value37, which is the chance ratio that an unmeasured confounder would wish to have with each the dependent and the impartial variable to utterly clarify away the noticed relationships. Unmeasured confounders with threat ratios of at the very least 1.77, 1.41, and a pair of.48 can be wanted to totally clarify the noticed relationships between Ct and end result, age and Ct, and pegivirus co-infection and Ct, respectively. In abstract, our analyses counsel that older people have increased viral masses and thus poorer outcomes, whereas these co-infected with pegivirus C have decrease viral masses and thus extra favorable outcomes.
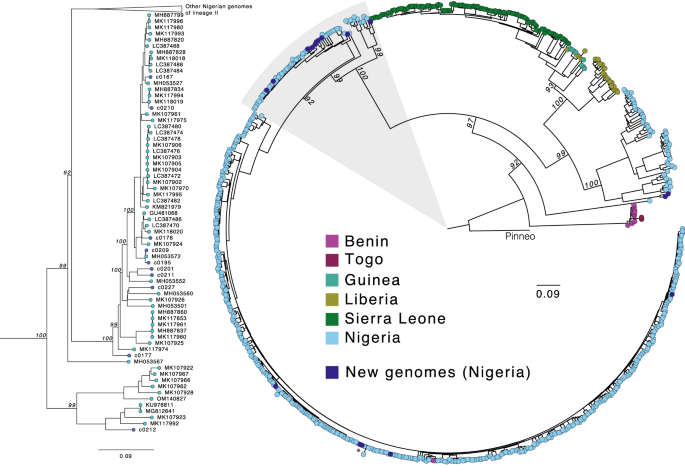
Subsequent, we additional investigated the genome sequences of a number of pathogens recognized within the LASV-positive and LASV-negative samples, starting with LASV itself, which is extremely genetically numerous. Its distinct viral lineages segregate geographically in Nigeria14, although most out there genome sequences are from the southwestern area. Our work generated 17 new high-quality (>90% of the genome assembled) LASV genomes, 15 from PCR-positive circumstances and two from PCR-negative circumstances. We noticed phylogenetic clustering of those samples by geographic origin, per earlier descriptions of geographic construction in LASV range in Nigeria (Fig. 3). Most of our genomes, together with these from the PCR-negative samples, had been of lineage II, and clustered in keeping with their sampling website (Irrua within the southwestern cluster and Ebonyi within the southeastern cluster). Two genomes from samples obtained in northwestern Nigeria clustered with lineage III genomes however fashioned a definite sub-clade, highlighting the extent of unsampled range on this poorly studied lineage.
Most probability phylogenetic tree of 17 new genomes (darkish blue) alongside 622 printed full S phase coding sequences. Ideas are coloured by the nation of pattern origin, and the tree is rooted within the Pinneo sequence (1979). The realm highlighted in grey, containing the vast majority of the brand new genomes (10/17), is proven in additional element on the left. The asterisk denotes the 2 RT-qPCR-negative samples that yielded full genomes. The dimensions bar denotes substitutions per website. Bootstrap values are proven on key nodes.
We additionally extra carefully examined our a number of hepatitis B, HIV-1, and pegivirus C genomes. All three hepatitis B genomes, from one LASV-positive and two LASV-negative people, had been categorised as subtype E, the predominant circulating genotype in Western and Central Africa38. A minimum of two of the seven HIV-1 genomes, from 4 LASV-positive and three LASV-negative samples, had been recombinant (Supplementary Desk 4). We constructed a phylogenetic tree with our 28 full pegivirus C genomes from 23 LASV-positive and 5 LASV-negative people and the opposite 130 annotated sequences out there in NCBI GenBank. The Nigerian genomes cluster with different African genomes, particularly these from Ghana and Cameroon, the closest nations represented within the tree (Supplementary Fig. 5).
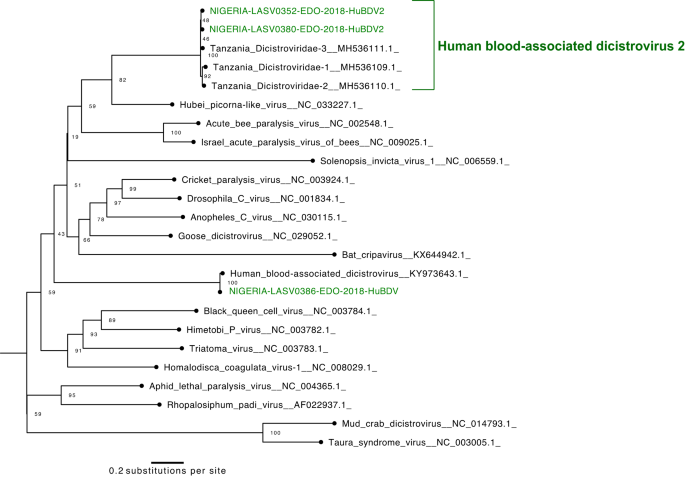
Lastly, we report the primary 4 Nigerian genomes of dicistroviruses, all of which had been present in LASV-positive samples. Dicistroviruses have primarily been described in arthropods39,40,41,42,43, although the poorly characterised human blood-associated dicistrovirus (HuBDV) was first found in a febrile Peruvian affected person in 201844. Right here, we assembled the second full HuBDV genome and one other partial genome. Furthermore, we assembled two extra unclassified dicistroviridae genomes, which had been >96% equivalent to sequences produced from febrile Tanzanian kids45 and extremely divergent from the HuBDV genomes (Fig. 4). We designate the clade that features our two unclassified genomes and the three Tanzanian genomes as human blood-associated dicistrovirus 2 (HuBDV-2; Fig. 4). Our identification of unlinked circumstances of HuBDV and HuBDV-2 means that these viruses could also be circulating extra broadly than recognized in Nigeria.
Cluster investigations yield genomic insights that inform public well being interventions
Genome sequencing has efficiently recognized the etiologies of illness outbreaks and decided the relationships between circumstances inside a cluster13,46,47,48. We investigated three separate outbreaks through the evaluation of 109 plasma samples collected by the NCDC. We examined all samples utilizing an RT-qPCR-based frequent pathogens panel (Supplementary Desk 5; Supplementary Information 1) and performed subsequent metagenomic sequencing on a subset of samples for outbreak characterization.
The primary cluster investigation consisted of 71 samples collected in 2017 from sufferers suspected to have mpox, brought on by monkeypox virus (MPXV). MPXV re-emerged in Nigeria over the identical calendar 12 months, after 40 years of absence, and sequencing of early circumstances steered spillover from a neighborhood reservoir, slightly than importation, because the supply49. Right here, we performed diagnostics and sequencing from plasma samples slightly than lesion swabs, that are heterogeneous samples that may be troublesome to gather from these with few or no seen lesions50. Although plasma is a extra standardized pattern kind, the diploma to which MPXV genetic materials is detectable in plasma is unknown. Of our 71 plasma samples, 35 had been optimistic for MPXV by qPCR (Supplementary Desk 6), indicating a minimal sensitivity of 49% for plasma testing (as not all sufferers had been sure to have MPXV). We chosen 5 MPXV-positive plasma samples—these with the best sequencing library quantification values—for unbiased sequencing in addition to hybrid seize with pan-viral goal enrichment probes (Strategies). Unbiased metagenomics yielded 30 or fewer aligned learn pairs for every pattern, whereas hybrid seize yielded as much as 20,000 aligned learn pairs (Supplementary Fig. 6). We produced contigs able to figuring out that the 5 samples belonged to the IIb clade (i.e., the clade answerable for the 2022 multinational outbreak), per different outbreak experiences49. We couldn’t assemble full genomes through both metagenomics or hybrid seize, probably due partly to the massive genome measurement, diminished viral masses within the blood relative to lesions51, and the Illumina MiSeq’s sequencing capability.
The second cluster investigation consisted of eight samples suspected to include yellow fever virus (YFV), collected in 2020 from Ebonyi, Edo, and Oyo states. YFV is the etiological agent of YF and in addition re-emerged in Nigeria in 2017 after a 40-year absence52. Beforehand, we reported YFV in a 2018 cluster with signs suggestive of LF and demonstrated that the circumstances had been extra carefully associated to modern Senegalese YFV genomes than to historic Nigerian sequences53. After confirming YFV was present in all eight samples through RT-qPCR, we sought to characterize the genomic ancestry of the 2020 outbreak. We produced two full YFV genomes, which belonged to the West Africa clade (Supplementary Fig. 7) and had been >98% just like sequences from the Nigerian 2018 YFV outbreak53, suggesting cryptic transmission and persistence of the 2018 YFV pressure. These knowledge contributed to the NCDC’s and World Well being Group’s (WHO) efforts to speed up vaccination campaigns and practice native healthcare employees within the analysis and remedy of YF54.
Lastly, we obtained 30 samples in November 2020 from a cluster in Benue, Nigeria, that offered with headache, diarrhea, vomiting, and stomach ache. The samples had been unfavorable for all pathogens within the RT-qPCR panel, and metagenomic sequencing of 12 samples did not establish an infectious etiology. The NCDC finally expanded its differential analysis to incorporate environmental causes, and the outbreak was decided to be as a result of pesticide poisoning55,56. Whereas metagenomics of a single pattern kind can not rule out an infectious trigger, this investigation emphasizes that it may assist public well being departments in updating their prior chances of particular diagnoses.
Metagenomics identifies viral infections in undiagnosed, extreme scientific circumstances
Within the scientific setting, metagenomic sequencing affords an alternative choice to the enumeration of single-pathogen diagnostic assessments, which might require a number of samples and finally be expensive and time-consuming57. Furthermore, in Nigeria and different LMIC settings, even massive hospitals at present solely have the capability to check for a small set of pathogens. We obtained eight plasma samples from people with scientific shows per an infectious etiology however with out proof of any generally circulating pathogens, collected in 2019–2020 from Ondo, Lagos, and Ebonyi states. Scientific and demographic metrics for these circumstances had been extremely different (Supplementary Desk 7).
We first screened the eight affected person samples towards the RT-qPCR frequent pathogens panel (Supplementary Desk 5; Supplementary Information 1) and did not establish any optimistic hits. Through unbiased metagenomic sequencing, we recognized viruses which are believable candidates for sickness in two sufferers. In a 3rd pattern, we detected Pegivirus C, a standard an infection in wholesome people58 that’s unlikely to be the reason for the scientific syndrome. No believable pathogenic viral taxa had been detected within the remaining 5 samples. Right here, we describe the scientific and genomic options of the circumstances with a putative analysis.
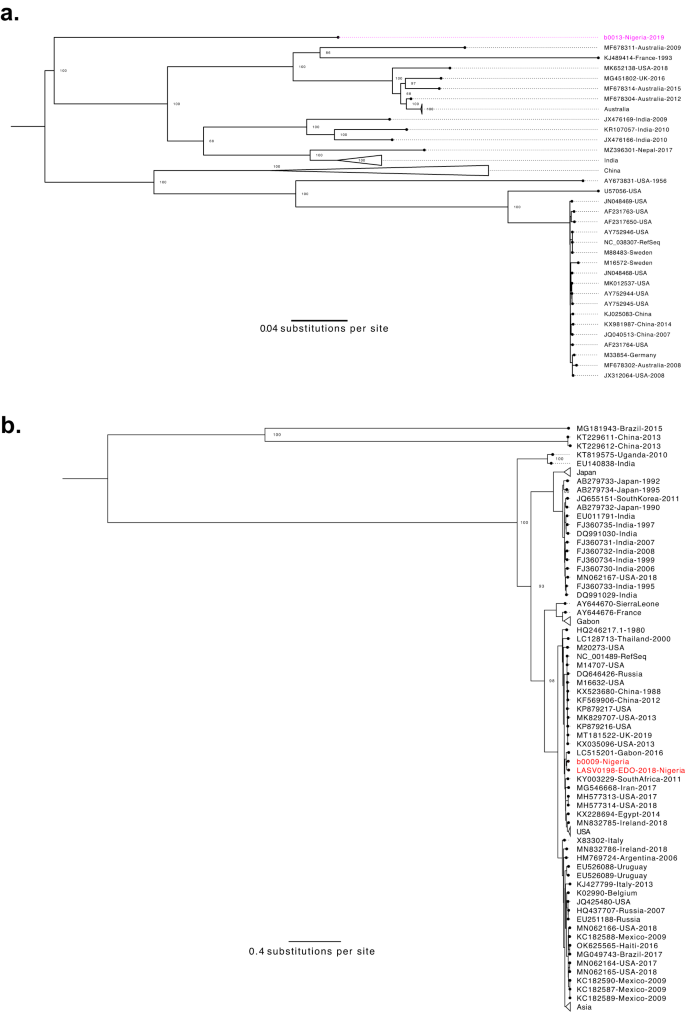
We recognized reads mapping to Enterovirus B within the plasma of a kid presenting with fever and seizures. We assembled a genome of Coxsackievirus-B3 (CV-B3; Fig. 5a), which is related to each gastrointestinal sickness and extra critical manifestations, together with myocarditis and meningitis59,60. The genome was most just like a CV-B3 genome from Japan (82% pairwise sequence identification), although the VP1 gene was most carefully associated to a partial genome from Nigeria (88% pairwise sequence identification to GQ496547.1)61.
a Coxsackievirus B3 (CV-B3) genetic range. Most probability phylogenetic tree with one new sequence (pink) alongside 63 full-length, printed sequences. Generated from whole-genome alignment (7447 bp). Bootstrap values for key nodes are proven. b Hepatovirus A genetic range. Most probability phylogenetic tree with two new sequences (crimson) alongside 105 full-length, printed sequences. Generated from whole-genome alignment (7736 bp). Bootstrap values for key nodes are proven.
We detected kind IB hepatovirus A (HAV; Fig. 5b) in one other little one presenting with left-sided weak spot, generalized lymphadenopathy, hepatosplenomegaly, and a head CT scan with proof of a proper hemispheric stroke. HAV, the causal agent of hepatitis A, is transmitted fecal-orally, sometimes presents with acute gastrointestinal manifestations, and barely causes demise62. This affected person’s signs usually are not per the textbook presentation of hepatitis A, although circumstances of neurological sequelae related to HAV have been documented63,64,65,66. We thus interpret the metagenomic sequencing outcomes with warning, as it’s potential that HAV is an incidental discovering. Nevertheless, we solely recognized HAV in 1 of our 592 different samples, suggesting that it’s an unusual co-infection and lending help to the chance that this affected person offered with an uncommon manifestation of HAV.