Cell tradition
Main human foreskin fibroblasts (HFF) have been established from the foreskin of a new child youngster in 1994 and have been used for analysis up to now6,7,14,15,16. Approval to make use of these cells for the research was obtained from the ethics committee of the medical council of Rheineland-Palatinate, Germany. HFF have been maintained in minimal important medium (MEM; Gibco-BRL, Glasgow, Scotland) supplemented with 5% fetal calf serum (FCS), 100 mg/l L-glutamine, 0.5 ng/ml fundamental fibroblast progress issue (bFGF, Invitrogen, Karlsruhe, Germany) and gentamicin (5 mg/l). For the experiments, HFF cell passage numbers between 16 to 19 have been used. HEC-LTT cells have been established by Dagmar Wirth and coworkers17. The cells have been derived from human umbilical vein endothelial cells (HUVECs) that have been conditionally immortalized with tetracycline-dependent expression of the SV40 large-T antigen and human telomerase reverse transcriptase (hTERT). For cultivation, tradition vessels have been coated with 0.1% gelatin (Sigma-Aldrich, Saint Louis, MO;) for a minimum of 30 minutes. HEC-LTT cells have been maintained in endothelial progress medium (EGM BulletKit; Lonza Gross sales Ltd., Basel, Switzerland) supplemented with 2 μg/mL doxycycline (Sigma-Aldrich, Saint Louis, MO). The proliferation of HEC-LTT cells may be managed by doxycycline (DOX). The addition of DOX prompts the expression of the immortalizing proteins hTERT and SV40 large-T antigen, leading to cell proliferation. Doxycycline was omitted throughout the complete experiments. Permission to make use of HEC-LTT cells for analysis functions was granted through a fabric switch settlement by the Helmholtz Centre for An infection Analysis (HZI). The cells have been proven to be permissive to HCMV an infection18. HEC-LTT have been kindly despatched to us by Christian Sinzger (Institute for Virology, Ulm College Medical Heart, Ulm, Germany) and used for experiments from passage 41 to passage 55.
Preparation of virus seed shares
Virus seed shares have been ready from supernatants of transfected HFF. Briefly, the pressure Towne-repΔGFP (hereafter denoted as TR-∆GFP) was reconstituted by transfection of bacterial synthetic chromosome (BAC) DNA, containing the Towne-repΔGFP genome into HFF. The technology of the BAC clone was described Lehmann et al.7. Transfected cells have been propagated till 100% of the cells confirmed cytopathic results (CPE). The virus-containing supernatants from these cultures have been harvested and precleared from mobile particles by centrifugation at 1,475 × g for 10 min after which saved as virus seed shares at −80 °C for additional propagation of the virus.
Technology of experimental shares
Virus experimental shares have been ready from supernatants of HFF, contaminated with virus seed shares. For this, HFF have been seeded at a density of 1.8 × 106 in 5 175 cm2 tissue tradition flasks and contaminated with 5 ml virus inoculum per flask. For this, 1 ml of the TR-∆GFP seed stock-supernatant and 4 ml of 5% MEM medium have been combined and added to the cells for 1.5 hours. Then 15 ml of contemporary 5% MEM medium was added and the contaminated cells have been incubated at 37 °C till the cultures confirmed an entire CPE. The cell tradition supernatants have been harvested and mixed. Mobile particles was eliminated by centrifugation at 1,475 × g for 10 min at room temperature. Lastly, the supernatants have been saved in freezing tubes at −80 °C.
Preparation of HCMV dense our bodies
For the purification of Dense Our bodies, twenty 175 cm2 tissue tradition flasks with 1,8 × 106 HFF have been contaminated with 1 ml of frozen virus supernatant shares of the HCMV pressure TR-ΔGFP, diluted in 4 ml 5% MEM medium. Following virus adsorption for 1.5 h, 15 ml of contemporary 5% MEM medium, supplemented with 50 nM of Letermovir (LMV) have been added and HFF have been incubated for a minimum of 7 days. LMV was added to the cell tradition media each 3 days after preliminary an infection. LMV is a extremely particular inhibitor of the HCMV terminase advanced and was proven to inhibit HCMV replication in cell tradition by interfering with the cleavage/packaging of HCMV genomes into nuclear capsids19. Supernatants from contaminated HFF that confirmed an entire cytopathogenic impact (CPE) have been harvested and gross mobile particles was eliminated by centrifugation for 10 min at 1,475 × g. Afterwards viral particles have been pelleted through ultracentrifugation at 95.000 × g for 70 min at 10 °C utilizing a 45Ti rotor in a Beckman Optima L-90K ultracentrifuge. For fractionation of the particles, the pellets have been resuspended in 2 ml of phosphate-buffered saline (PBS) and loaded onto glycerol-tartrate density gradients. For gradient preparation, 5 ml of a 35% Na-tartrate answer in 0.04 M Na-phosphate buffer, pH 7.4 and 4 ml of a 15% Na-tartrate–30% glycerol answer in 0.04 M Na-phosphate buffer, pH 7.4 have been combined in a gradient mixer and launched right into a polycarbonate centrifuge tube (14 ml; Beckman Extremely-Clear centrifuge tubes) at an angle of 45°. Gradients have been overlaid with 1 ml of the concentrated viral particle suspension and centrifuged in a Beckman SW41Ti swing-out rotor for 60 min at 90,000 × g and 10 °C with out deceleration. Subsequently, the DB-fraction was visualized by gentle scattering and picked up by puncturing the tube with a syringe. The fraction collected from gradients containing DB have been washed with 10 ml PBS and DB have been concentrated by ultracentrifugation utilizing a SW41Ti swing-out rotor for 90 min at 98,000 × g and 10 °C. Lastly, the DB-pellet was resuspended in 250 µl PBS. Aliquots of 30 µl have been ready and saved at −80 °C till additional use. For the dedication of DB-protein concentrations, the Pierce™ BCA Protein Assay Equipment (23225, ThermoFisher Scientific, Darmstadt, Germany) was used in accordance the producers protocol.
DB have been thawed and irradiated with ultra-violet (UV) gentle shortly earlier than they have been utilized to cells. Following resuspension in a complete quantity of 120 µl PBS, DB have been transferred onto a spot plate and UV-irradiated at a wavelength of 254 nm for two minutes. Then, 100 µl of the UV-irradiated DB/PBS answer have been combined with 2,9 ml tradition medium and added to the cells.
Preparation of protein extracts for proteomic evaluation
HFF have been seeded at a density of 0.5 × 106 in two 10 cm dishes. On the subsequent day, 20 µg of TR-∆GFP- derived DB have been UV-irradiated and added to every dish. The DB-inoculum was incubated for two h. Afterwards, 7 ml MEM medium was added and cells have been incubated for extra 22 h. Subsequent, the medium was eliminated and the cells have been washed twice with PBS. The HFF of two dishes have been pooled and the cell quantity was decided. 1 × 106 fibroblasts have been lysed in 40 µl 2x Laemmli buffer with out bromophenol-blue staining and heated at 99 °C for 10 min. After cooling, NuPAGE LDS Pattern Buffer (4x) (Life applied sciences) and 100 mM DTT have been added and the samples have been incubated at 70 °C for additional 10 min.
Endothelial cells have been seeded at a density of 0.6 × 106 in two 10 cm dishes in absence of doxycycline. ECs have been uncovered to 40 µg of UV-irradiated DB of the HCMV pressure TR-∆GFP. The next steps have been carried out as described for HFF.
Protein in-gel digestion
Proteins have been loaded onto a ten% NuPAGE Bis-Tris gel and resolved briefly. Following that, the gel was stained with Coomassie blue and reduce into small cubes utilizing a clear scalpel. Gel destaining was carried out in 50% ethanol/25 mM ammonium bicarbonate. Protein discount was carried out in 10 mM DTT at 56 °C, adopted by alkylation in 50 mM iodoacetamide at midnight at room temperature. Trypsin (1 µg per pattern) was used to digest the proteins in 50 mM TEAB (triethylammonium bicarbonate) buffer in a single day at 37 °C. Peptide extraction was carried out sequentially in 30% and 100% acetonitrile. Thereafter, the pattern quantity was diminished in a centrifugal evaporator to take away residual acetonitrile. Then, the pattern was stuffed with 100 mM TEAB to succeed in a last pattern quantity of 100 µl.
Dimethyl-labelling
In keeping with the experimental design scheme20, the digested samples have been labelled as “Gentle”, “Medium” or “Heavy” by including 4 µl of 4% formaldehyde, formaldehyde-d2 or formaldehyde-13C, d2 answer, respectively. This was then adopted by addition of 4 µl of 0.6 M NaBH3CN (to “Gentle” or “Medium” pattern) or NaBD3CN (to “Heavy” pattern). Thereafter, the samples have been incubated at room temperature with orbital shaking for 1 h. The labelling response was then quenched by including 19 µl of 1 M ammonium bicarbonate (last focus 150 mM) and incubated at room temperature with orbital shaking for 15 min. Afterwards, peptides have been acidified with formic acid to succeed in pH ~3. The paired labelled samples have been then mixed. The resultant peptide answer was purified by stable part extraction in C18 StageTips21).
Lqiuid chromatography tandem mass spectrometry
Peptides have been separated in an in-house packed 30-cm analytical column (inside diameter: 75 μm; ReproSil-Pur 120 C18-AQ 1.9-μm beads, Dr. Maisch GmbH; heated at 40 °C) by on-line reversed part chromatography by way of a 225-min non-linear gradient of 1.6–32% acetonitrile with 0.1% formic acid at a nanoflow fee of 225 nl/min. The eluted peptides have been sprayed straight by electrospray ionization right into a Q Exactive Plus Orbitrap mass spectrometer (Thermo Scientific). Information-dependent acquisition was carried out utilizing a top10 methodology. Following every full scan (mass vary: 300 to 1,650 m/z; decision: 70,000, goal worth: 3 × 106, most injection time: 20 ms), as much as 10 MS2 scans have been carried out through larger vitality collision dissociation (normalised collision vitality: 25%, decision: 17,500, goal worth: 1 × 105, most injection time: 120 ms, isolation window: 1.8 m/z). Cost state choice was carried out by rejecting precursor ions of unassigned or +1 cost state. Dynamic exclusion time was set to 35 s.
Mass spectrometry information processing and statistical evaluation
Uncooked information information have been processed by MaxQuant software program bundle (model 2.1.3.0)22 utilizing Andromeda search engine23. Spectral information have been searched towards a target-decoy database consisting of the ahead and reverse sequences of UniProt proteomes downloaded on 10th August 2022 listed for the particular Taxon IDs (ID 9606, H. sapiens, 79759 entries; ID 10359, HCMV, 17993 entries; ID 10363, HCMV City pressure, 304 entries) and a listing of 246 frequent contaminants. Corresponding dimethyl labels have been assigned as “Gentle” (DimethLys0 and DimethNter0), “Medium” (DimethLys4 and DimethNter4) and “Heavy” (DimethLys8 and DimethNter8) based on the labelling scheme. For every peptide, as much as 3 labelled amino acids have been allowed. Trypsin/P was chosen for enzyme specificity. Carbamidomethylation of cysteine was chosen as mounted modification. Protein N-terminus acetylation and oxidation of methionine have been assigned in variable modifications. As much as 2 missed cleavages have been tolerated. A minimal peptide size of seven amino acids was required. For each peptide and protein identifications, a false discovery fee (FDR) of 1% was chosen.
For protein quantification, minimal ratio depend was set to at least one. Each the distinctive and razor peptides have been used for quantification. The “re-quantify” perform was switched on. The “superior ratio estimation” possibility was additionally chosen. Reverse hits and potential contaminants have been filtered out. Protein teams with a minimum of one distinctive peptide have been retained. Ratios of label-swapped samples have been inverted to symbolize dense our bodies remedy (DB) over management for all technical and organic replicates. The normalized ratios have been then log2 reworked and median-centered. Afterwards, the ratios of the technical replicates belonging to the identical organic replicate have been averaged ignoring the lacking worth if current.
Following the above-mentioned steps, statistical evaluation to determine differentially-regulated proteins was carried out utilizing the limma software program bundle in R24. For fibroblasts, proteins with ratios in a minimum of three out of 5 organic replicates have been retained. For endothelial cells, proteins with ratios in a minimum of two organic replicates have been retained. A linear mannequin was then fitted to evaluate the ratios for every protein with out additional adjustment for a number of testing. The log2 fold change and the importance of the distinction have been displayed in a volcano plot. Solely proteins with a minimal log2 fold change of 1 and a p worth decrease than 0.05 have been thought of as being differentially regulated.
Proteome evaluation of fibroblasts uncovered to dense our bodies
In our proteomics research, we investigated the impression of HCMV Dense Our bodies (DB) incubation on two totally different cell varieties. The adjustments within the mobile proteome of fibroblast or endothelial cells upon DB software was in comparison with mock-treated reference samples.
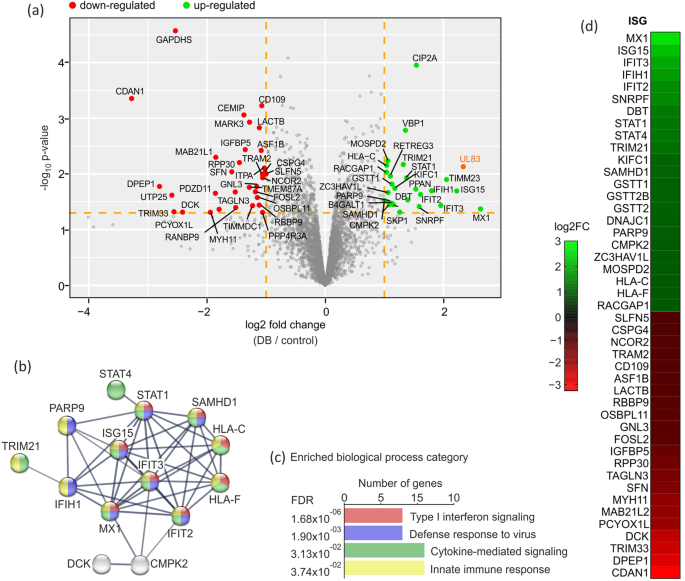
Involving 5 organic replicates, 3757 protein teams have been recognized in complete in fibroblasts after a number of steps of stringency filtering and limma evaluation. Utilizing a twofold cut-off, 153 proteins remained, of which 68 confirmed a p worth decrease than 0.05. These 68 proteins have been thought of as being differentially expressed at 24 h submit DB-application to fibroblasts and are listed in Desk 1. Outcomes are displayed in a volcano plot in Fig. 1a. 33 proteins have been upregulated whereas 35 have been downregulated in DB-treated cells compared to mock-treated cells. To characterize the relationships between the 68 differentially expressed proteins, the net device STRING (http://string-db.org/, accessed on 16.05.2023) was utilized to analyse the interacting companions. The protein-protein interplay (PPI) community evaluation, depicted in Fig. 1b reveals one foremost cluster composed of proteins that perform in organic processes of kind I interferon response, protection response to virus, response to virus and cytokine-mediated signalling pathways (Fig. 1b). The bar chart in Fig. 1c reveals the enriched organic processes organized based on rising False Discovery Charges (FDR). Strikingly, when the 68 altered proteins have been submitted to the Interferome database on-line device (v2.01, accessed on November 202225), the bulk (66%) of them have been recognized as being interferon-stimulated genes (ISGs) (Fig. 1d).
Proteome evaluation of DB-regulated proteins in HFF. HFF have been mock-treated or incubated with 20 µg of UV-inactivated DB derived from TR-∆GFP. Cell lysates have been subjected to complete proteome MS/MS evaluation at 24 h submit DB- software. (a) Volcano plot exhibiting the log2 fold-change (x-axis) versus the importance (y-axis) of the in complete 3757 proteins, detected in 5 organic replicates. The dotted strains in orange present the cut-off fold change of ±1.0 and a p-value of 0.05. The importance (non-adjusted p-value) and the fold-change are transformed to −log10(p-value) and log2 fold-change, respectively. There have been 33 proteins elevated by >1.0-fold with p-value < 0.05 (inexperienced dots), and 35 proteins that have been decreased by < − 1.0-fold with p-value < 0.05, (purple dots). The HCMV tegument protein pp65 (UL83) is highlighted in orange and its detection was used as a constructive management, indicating DB internalisation into HFF. The volcano plot was generated utilizing the R software program. (b + c) Protein-Protein Interplay (PPI) evaluation and useful classification of the regulated proteins in HFF after DB-stimulation. (b) Show of the STRING PPI community generated upon getting into the 68 regulated proteins into the STRING database. The community nodes symbolize all of the proteins produced by a single protein-coding gene locus. Nodes are colored based on their perform within the indicated organic processes in c. Gray nodes point out proteins linked to the enter proteins however with out affiliation with the organic processes. Connections mirror protein interplay and the road thickness signifies power of the information help, utilizing a excessive confidence cut-off with a rating of 0.7. Proteins with no interplay to different proteins within the community have been eliminated. (c) Bar chart of the organic processes, linked to the proteins that have been discovered to be regulated in HFF after DB-stimulation. The association was carried out based on rising False Discovery Charges (FDR). The y-axis represents organic course of classes, whereas the x-axis signifies the variety of genes concerned in every class. (d) Heatmap of the 45 altered ISGs. The expression patterns have been organized hierarchically based mostly on the imply of the log2 transformed normalized ratio from 5 organic replicates. The log2FC is represented with a color gradient. IFN, Interferon; ISG, Interferon-stimulated gene; STRING, Search Device for the Retrieval of Interacting Genes.
Proteome evaluation of endothelial cells uncovered to Dense Our bodies
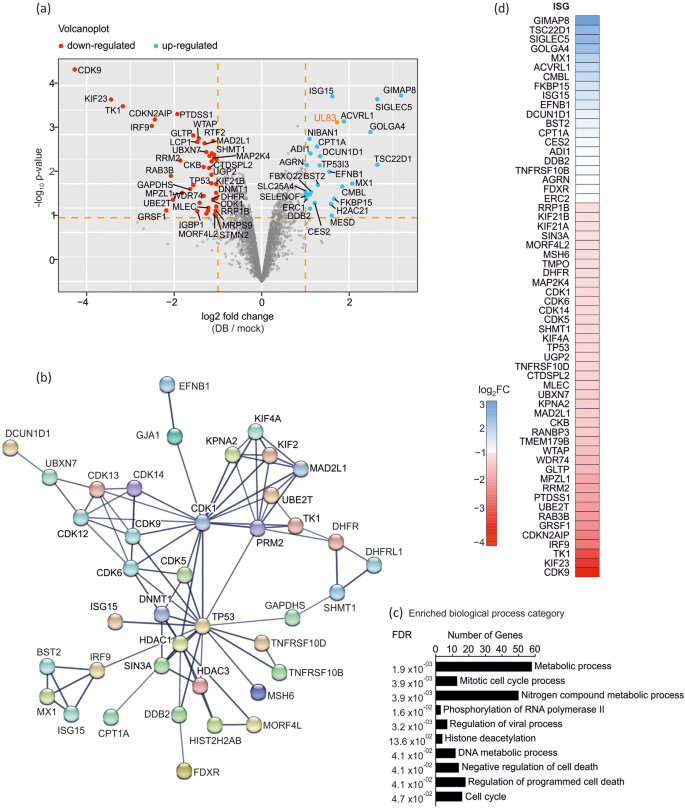
Mobile proteins regulated upon publicity of Dense Our bodies to endothelial cells have been analysed from a minimum of two organic replicates. On the idea of the filtering standards beforehand used, 83 altered proteins (listed in Desk 2) have been discovered to be differentially regulated and are proven within the volcano plot in Fig. 2a. To determine the results of DB-treatment on mobile pathways, the STRING database (https://string-db.org, accessed on 20.10.2022) was used. The PPI community in Fig. 2b reveals the interactions between the 83 regulated proteins, utilizing the excessive confidence interplay rating of 0.7. Every of the differentially expressed proteins mapped to 3 or 4 main useful networks that have been linked by the hub proteins CDK1 or TP53. One cluster consists of the proteins MX1, ISG15, BST2 and IRF3 that are identified to perform within the kind I interferon signalling pathway. The opposite two strongly linked networks comprise CDKs and KIF- proteins, each related to the cell cycle. The highest ten classes of organic processes that have been enriched upon DB-application in endothelial cells are depicted within the bar chart in Fig. 2c and are organized based on rising False Discovery Charges (FDR). The 83 proteins have been submitted to the Interferome database (v2.01, accessed November 2022)25. 60 proteins have been recognized as interferon stimulated genes, most of which have been downregulated (Fig. 2nd).
Quantitative proteomic evaluation of differentially regulated proteins in DB-treated ECs.. Endothelial cells have been incubated with 40 µg of UV-inactivated DB (pressure TR-∆GFP) or left untreated. Cell lysates have been ready 24 h after software and subjected to proteome evaluation. (a) The 2719 proteins recognized by MS from three organic replicates are proven in a volcano plot based on their statistical p-value (y-axis) and their relative abundance ratio (log2 fold change) between DB- and mock-treated cells (x-axis). Pink dots point out differentially expressed proteins that have been considerably downregulated after DB remedy (fold change > 1.0; p < 0.05). Blue dots point out differentially expressed proteins that have been considerably upregulated (fold change < 1.0; p < 0.05). The viral tegument protein pp65 (UL83) is highlighted in orange and was used as a management for DB internalisation into ECs. The volcano plot was generated utilizing the R software program. (b) STRING Protein-Protein Interplay community of the 83 proteins that have been differentially expressed in ECs upon DB remedy. Proteins with no associations to different proteins within the community have been eliminated. Community nodes symbolize all of the proteins produced by a single, protein-coding gene locus. Traces depict protein interplay and the road thickness signifies the power of the information help with a minimal confidence cut-off of 0.7 (excessive confidence). (c) Bar chart of the enriched organic processes related to differentially expressed proteins. The highest ten enriched organic processes are organized based on rising False Discovery Charges (FDR). The y-axis represents organic course of classes, whereas the x-axis signifies the variety of genes concerned in every class. (d) 60 differentially regulated proteins have been designated as IRGs. The log2FC is represented with a color gradient. The 20 up-regulated ISGs are indicated in blue and the 40 down-regulated ISGs are indicated in purple. STRING, Search Device for the Retrieval of Interacting Genes.
The validation of the MS-analyses supplied right here was printed elsewhere11. In that research, we evaluated the expression of the chosen proteins MX1, IFIT3 and ISG15 in fibroblasts and endothelial cells utilizing Western blot analyses. The Western blot outcomes have been in step with the outcomes obtained from the MS information. A strong enhance within the expression ranges of all three proteins upon the DB-treatment could possibly be confirmed. Though the fold adjustments weren’t similar within the immunoblot analyses, in comparison with the MS information at 24 h.p.a., the tendencies have been related. Taken collectively, these experimental outcomes present that our proteomics information are dependable.