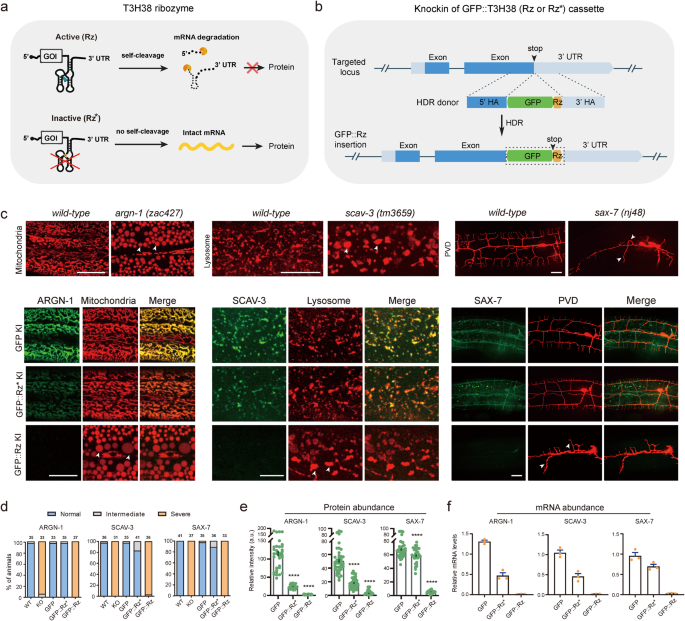
T3H38 ribozyme effectively cis-cleaves endogenous mRNAs in C. elegans
To check whether or not the T3H38 ribozyme is adaptable for C. elegans, we envisioned that CRISPR/Cas9-mediated knock-in of the energetic T3H38 ribozyme into the three’ untranslated area (UTR) of an endogenous gene would result in ribozyme-mediated self-cleavage and mRNA degradation because the mRNA doesn’t comprise a 3’ UTR13. In distinction, using a catalytically inactive mutant kind would generate a stabilized mRNA and subsequently information regular protein manufacturing (Fig. 1a). Towards this objective, we inserted the coding sequence for the inexperienced fluorescence protein (gfp), gfp-inactive T3H38 (gfp-Rz*), or gfp-active T3H38 (gfp-Rz) into the instantly upstream of the three’ UTR of three endogenous genes, respectively (Supplementary Fig. S1)10. These genes are argn-1 (a gene required for regular mitochondrial morphology), scav-3 (a gene required for regular lysosomal morphology) and sax-7 (a gene required for correct dendrite branching and steerage) (Fig. 1b)14,15,16,17. Within the three strains inserted with C-terminal gfp, the morphologies of mitochondria, lysosomes or the PVD dendrites have been just like that of wild-type management, suggesting that tagging these genes with a C-terminal gfp doesn’t have an effect on their features (Fig. 1c, d). Subsequent, we examined the three strains during which the goal genes have been tagged with gfp-inactive T3H38. We discovered that 97%, 83% and 89% animals confirmed regular morphologies of mitochondria, lysosomes, and dendrites, respectively (Fig. 1c, d). Notably, quantitative fluorescent imaging revealed that the depth of GFP-Rz*-fused ARGN-1 and SCAV-3 proteins have been downregulated by 79% and 64%, respectively, whereas the extent of GFP-Rz*-fused SAX-7 protein was solely barely lowered (Fig. 1e). These outcomes counsel that putting the inactive ribozyme within the 3’ UTR could have an effect on the expression of some goal genes, presumably as a consequence of varied components similar to defects in mRNA stability, post-transcriptional modification, transport or translation. Nevertheless, the remaining proteins of ARGN-1 and SCAV-3 seemed to be adequate to take care of the traditional morphology of organelles. We then examined the three strains carrying gfp-active T3H38 insertions. As anticipated, all of them exhibited phenotypes just like robust loss-of-function or null mutants14,15,16,17, suggesting that the endogenous gene expression was virtually fully suppressed, which is supported by our quantification of the GFP depth (Fig. 1d, e). Quantitative reverse transcription PCR (qRT-PCR) analyses have been according to the notion that energetic T3H38 effectively cleaved goal mRNAs in all strains (Fig. 1f).
a Schematic of energetic T3H38 ribozyme (Rz) for inducing an off-switch of gene expression. Tailored with permission from Wurmthaler et al.11. An inactive T3H38 (Rz*) was generated as a management. b Schematic diagram of CRISPR/Cas9-mediated homologous recombination to combine GFP, GFP::Rz or GFP::Rz* on the C-terminal of endogenous loci. c Confocal fluorescent photos to point out the morphology of mitochondria (Pcol-19::mito-mKate), lysosomes (Pced-1::nuc-1::mCherry) and PVD (ser2prom::myr-mCherry) in associated to the protein expression stage of ARGN-1::GFP, SCAV-3::GFP and SAX-7::GFP, respectively, underneath completely different circumstances. 3-day-old adults have been imaged for the argn-1 and scav-3 teams; 2-day-old adults have been imaged for the sax-7 group. Scale bars: 20 μm. Arrowheads: enlarged mitochondria/lysosome or dendrite branching defects. argn-1(zac427), scav-3(tm3659) and sax-7(nj48) are deletion mutants, leading to robust loss-of-function/null alleles, and are used for comparisons. d Quantifications of the proportion of animals containing regular, intermediately faulty and severely faulty morphology of mitochondria (argn-1 group), lysosomes (scav-3 group) and PVD dendrites (sax-7 group), respectively. The variety of animals quantified for every group was indicated above the columns. e Expression of ARGN-1::GFP, SCAV-3::GFP and SAX-7::GFP, as proven in (c), have been quantified primarily based on fluorescent imaging. Information are displayed as imply ± s.e.m. Every dot represents a single worm. Variety of animals quantified: n = 35 for ARGN-1::GFP; n = 37 for ARGN-1::GFP::Rz*; n = 31 for ARGN-1::GFP::Rz; n = 41 for SCAV-3::GFP; n = 40 for SCAV-3::GFP::Rz*; n = 34 for SCAV-3::GFP::Rz; n = 41 for SAX-7::GFP; n = 32 for SAX-7::GFP::Rz*; n = 36 for SAX-7::GFP::Rz. ****p < 0.0001 (one-way ANOVA with the Tukey correction). f Relative mRNA abundance of goal genes at completely different circumstances in (c) measured by quantitative RT-PCR. Three organic replicates have been quantified and proven. All values are displayed as imply ± s.e.m. Supply information are offered as a Supply Information file.
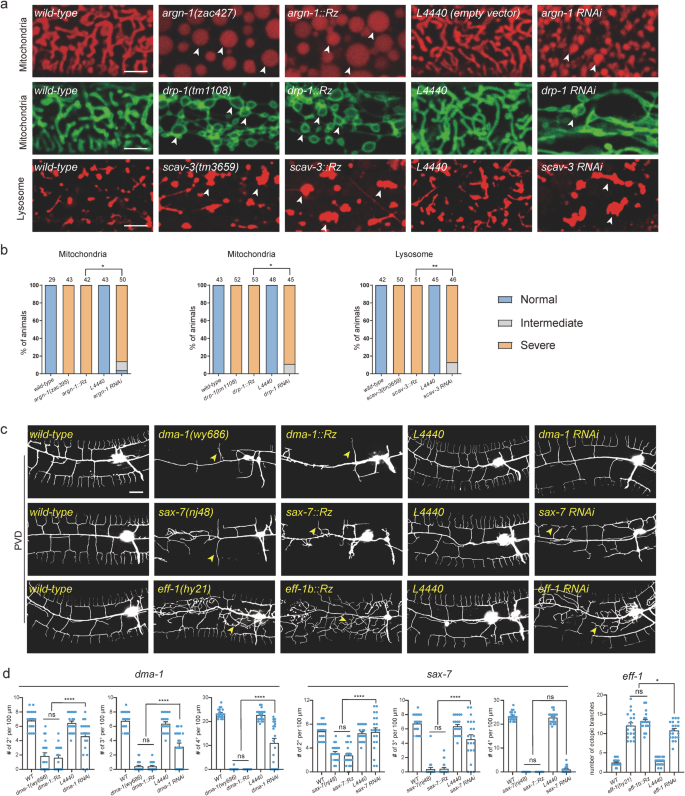
To additional exhibit the efficacy and flexibility of ribozyme-mediated gene regulation, we prolonged our investigation to incorporate a broader vary of genes. Particularly, we inserted loxP-T3H38-loxP into 5 extra genes, together with dhgd-1, hphd-1, drp-1, mnr-1 and eff-1, that are important for sustaining regular mitochondrial morphology or PVD dendrite arborization16,18,19,20,21. Our T3H38 ribozyme-based technique achieved strong knockdown of endogenous protein expression for all these genes, as evidenced by the null-like loss-of-function phenotypes displayed by the T3H38 knock-in alleles (Supplementary Fig. S2a–c).
The above eight genes act within the dermis to regulate correct morphologies of mitochondria and lysosomes inside the dermis and PVD dendrites14,15,16,17,18,19,20,21. To check whether or not the T3H38 ribozyme is efficient in different varieties of tissues, we inserted the sequence of T3H38 into the endogenous locus of unc-86, mec-3, dma-1, hpo-30, kpc-1, rab-10 and lect-2, which perform within the PVD neurons (for the primary six genes) and physique wall muscular tissues (for lect-2), respectively, to regulate dendrite branching22,23,24,25,26,27,28,29. Our outcomes point out that every one ribozyme knock-in strains, besides kpc-1-loxP-ribozyme-loxP, confirmed virtually equivalent phenotypes because the genetic null mutant animals, suggesting that T3H38 efficiently blocks the endogenous expression of those six genes in neurons and muscular tissues, respectively (Supplementary Fig. S2d, e). Nevertheless, inserting the ribozyme into the kpc-1 locus didn’t lead to a loss-of-function phenotype, suggesting that the self-cleavage exercise is perhaps affected when T3H38 was inserted into some particular uncommon sequence context. Collectively, these outcomes exhibit that T3H38 ribozyme effectively eliminates endogenous gene expression in a number of varieties of tissues when inserted into the 5’ of the three’ UTR for a lot of the genes we examined in C. elegans. We named this ribozyme-mediated inactivation of endogenous gene expression as Ribo-Off. Thus, strong gene knockout will be achieved with a single genetic modification by inserting the 63 bp T3H38 ribozyme sequence into the three’ UTR of a gene of curiosity.
We subsequent in contrast the knockdown effectivity of Ribo-Off and RNA interference for six genes primarily based on the severity of their loss-of-function phenotypes. Our outcomes confirmed that inserting the ribozyme into argn-1, drp-1 and scav-3 led to virtually 100% of animals displaying severely enlarged mitochondria or lysosome phenotypes, whereas gene-specific dsRNAs resulted in intermediate phenotypes in small however vital proportions of animals (Fig. 2a, b). Lack of endogenous eff-1 brought about ectopic branching within the lateral areas, whereas lack of dma-1 or sax-7 resulted in lowered higher-order department formation (tertiary and quaternary branches)16,20,22. The entire-body Ribo-Off alleles of eff-1, dma-1 and sax-7 phenocopied the beforehand reported robust lack of perform or null alleles, whereas eff-1 (RNAi), dma-1(RNAi) and sax-7 (RNAi) animals confirmed much less extreme dendrite branching defects (Fig. 2c, d). Total, our findings counsel that Ribo-Off outperformed RNAi, a minimum of for the six genes we examined.
a Confocal photos of C. elegans expressing transgenic markers to label mitochondria or lysosomes in wild-type and mutant animals. Mitochondria have been labeled through zjuSi47[Pcol-19>mito::mKate2] or zjuSi48[Pcol-19>tomm-20::gfp]. Lysosomes have been labeled through zacSi13(Psemo-1>ctns-1-mcherry). Arrowheads: abnormally enlarged mitochondria or lysosomes. Scale bars: 5 μm. b Quantification of organelle morphology phenotypes. *p < 0.05; **p < 0.01, χ2 check. The variety of animals quantified for every group was indicated above the columns. c Confocal photos displaying dendrite morphologies in wild-type and mutant animals. PVD dendrites have been labeled through wyIs592[PVD>myr-gfp]. Scale bars: 20 μm. d Quantifications of dendrite branching phenotypes in wild-type and mutant animals. All values are introduced as imply ± s.e.m. For sax-7(nj48) and sax-7::Rz: n = 21 animals; For all different genotypes: n = 20 animals. ns: not vital. *p < 0.1. ****p < 0.0001 (one-way ANOVA with the Tukey correction). Supply information are offered as a Supply Information file.
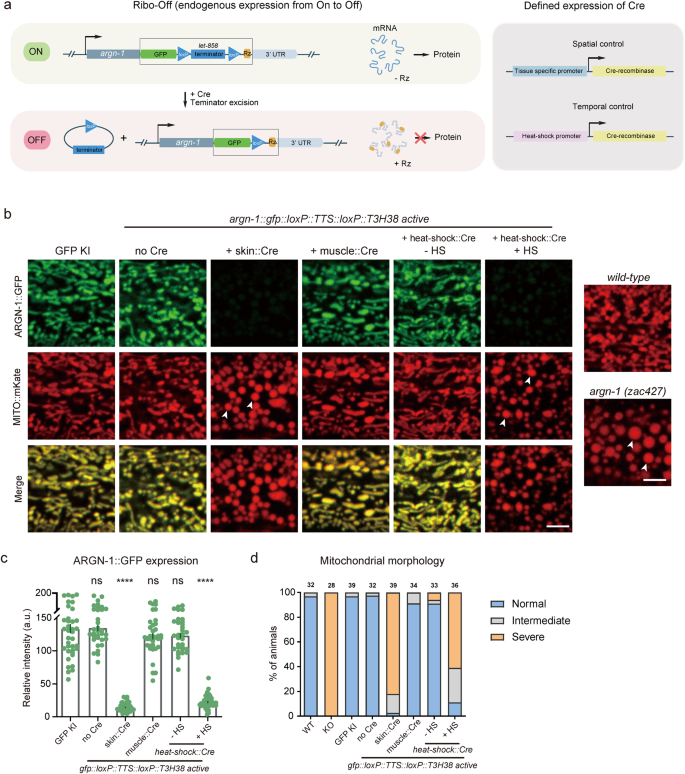
Flip off endogenous gene expression in area and time through Ribo-Off
Inspired by the whole-body Ribo-Off described above, we sought to make use of ribozyme to carry out conditional gene knockout. To realize this function, we used CRISPR/Cas9 know-how to insert gfp::loxP::let-858 transcriptional termination sequence (TTS)::loxP::T3H38 ribozyme instantly after the cease codons of the abovementioned three genes, argn-1, scav-3 and sax-730,31. Theoretically, the endogenous genes can use the let-858 3’ UTR to stabilize the mRNAs with out the expression of the recombinase Cre protein. Nevertheless, when Cre is expressed in a particular tissue (through tissue-specific promoters) or at a particular developmental stage (through the heat-shock promoter), the loxP- TTS-loxP cassette might be deleted from the endogenous loci, and the mRNAs might be unstable and degraded as a consequence of T3H38-mediated self-cleavage, thus leading to conditional knockout of goal genes (Fig. 3a).
a Schematics to point out the designs for attaining spatiotemporal management of Cre expression (left) and the inducible T3H38 ribozyme/Cre-loxp system to conditionally knockdown endogenous expression. b Confocal fluorescent photos to point out the morphology of mitochondria labeled by MITO::mKate and ARGN-1::GFP expression ranges underneath completely different circumstances. 2-day-old grownup animals have been imaged. Scale bars: 5 μm. Arrowheads: enlarged mitochondria. c Relative expression of ARGN-1::GFP underneath varied circumstances as proven in (b). All values are introduced as imply ± s.e.m. Every dot represents a single animal. n = 40 for ARGN-1::GFP; n = 33 for ARGN-1::loxP::TTS::loxP::Rz with out Cre; n = 43 for ARGN-1::loxP::TTS::loxP::Rz with Pores and skin Cre; n = 36 for ARGN-1::loxP::TTS::loxP::Rz with muscle Cre; n = 32 for ARGN-1::loxP::TTS::loxP::Rz with heat-shock Cre with out heat-shock; n = 39 for ARGN-1::loxP::TTS::loxP::Rz with heat-shock Cre with heat-shock. ****p < 0.0001 (one-way ANOVA with the Tukey correction). d Quantifications of the proportion of animals containing regular, intermediately faulty and severely faulty morphology of mitochondria. The variety of animals quantified for every group was indicated above every column. Supply information are offered as a Supply Information file.
Certainly, all of the strains carrying gfp::loxP::TTS::loxP::T3H38 ribozyme knock-in have been indistinguishable from the wild-type and the gfp knock-in management teams for the morphologies of mitochondria, lysosomes and dendrites, respectively (Fig. 3b and Supplementary Fig. S3a, b). Particular expression of Cre within the dermis considerably lowered protein expression and generated faulty morphologies, just like that of the whole-body knockout animals, suggesting that the features of those genes within the dermis are dramatically impaired (Fig. 3b and Supplementary Fig. S3a, b). As a management, expressing the Cre within the muscle cells neither affected the expression of those genes nor generated any loss-of-function phenotypes within the dermis, demonstrating that the conditional knockout is restricted (Fig. 3b and Supplementary Fig. S3a, b). To check whether or not we might regulate the expression of endogenous genes in a temporal method, we used a heat-shock promoter to drive Cre expression at particular time factors. When the animals weren’t handled with heat-shock, mitochondrial or lysosomal morphology was regular. In distinction, warmth shock for 1 h and restoration for 48 h virtually completely blocked the expression of those genes and generated faulty phenotypes just like that of whole-body knockout management animals (Fig. 3c, d and Supplementary Fig. S3c–f).
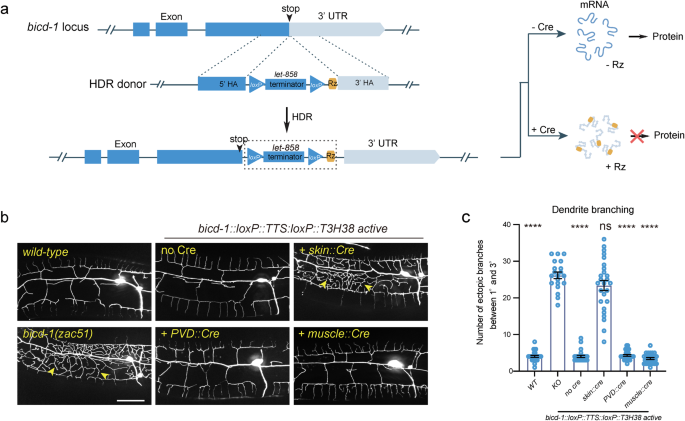
Ribo-Off can flip off important genes in particular tissues
Subsequent, we sought to find out whether or not our ribozyme-based conditional knockout technique applies to important genes. We inserted the loxP::TTS::loxP::ribozyme sequence instantly after the cease codons of bicd-1, a necessary gene required for correct dendrite morphogenesis (Fig. 4a). bicd-1 null mutants are embryonic deadly32. Curiously, we remoted bicd-1(zac51) from a ahead genetic display screen for dendrite morphogenesis irregular mutants, which confirmed an ectopic department formation defect just like the bicd-1 (RNAi) animals32. The mutant animals have been viable, probably as a result of zac51 is a partial loss-of-function allele. This pressure was used as a constructive management. We discovered that the bicd-1::loxP::TTS::loxP::ribozyme genetically modified animals developed and generated progenies usually with out Cre expression. A earlier research has reported that bicd-1 acts in PVD neurons to regulate dendrite branching32. Nevertheless, utilizing our newly developed ribozyme-based conditional Ribo-Off method, we discovered that knockout bicd-1 within the PVD cell lineage or the muscular tissues didn’t generate a dendrite branching defect, whereas inactivation of bicd-1 in dermis phenocopied the loss-of-function phenotype, demonstrating a non-cell autonomous perform of BICD-1 controlling dendrite morphogenesis (Fig. 4b, c). Thus, by combining the ribozyme and Cre/LoxP methods, we have been in a position to inactivate the expression of the endogenous genes in each area and time.
a Schematic illustration of utilizing CRISPR/Cas9-mediated homologous recombination to insert the GFP::loxP::TTS::loxP::Rz cassette to the C-terminal of the endogenous bicd-1 locus, and the associated final result of protein expression with or with out Cre expression. b Confocal fluorescent photos to point out the morphology of PVD dendrites. Cre recombinase was particularly expressed to delete the let-858 transcriptional terminator in varied tissues utilizing completely different tissue-specific promoters. L4-stage animals with the heat-shock::Cre transgene have been handled with heat-shock. 3-day-old grownup worms have been imaged for all teams. Scale bars: 20 μm. c Quantifications of variety of ectopic dendritic branches between the first dendrites and the tertiary dendrites in a area 100 µm anterior to the PVD cell physique in several genetic backgrounds as proven in (b). All values are introduced as imply ± s.e.m. Variety of animals quantified: n = 27 for WT; n = 20 for bicd-1(zac51); n = 26 for bicd-1::loxP::TTS::loxP::Rz with out Cre; n = 26 for bicd-1::loxP::TTS::loxP::Rz with pores and skin Cre; n = 28 for bicd-1::loxP::TTS::loxP::Rz with PVD Cre; n = 27 for bicd-1::loxP::TTS::loxP::Rz with muscle Cre. ns: not vital. ****p < 0.0001 (one-way ANOVA with the Tukey correction). Supply information are offered as a Supply Information file.
Activate endogenous gene expression through morpholino-regulated T3H38
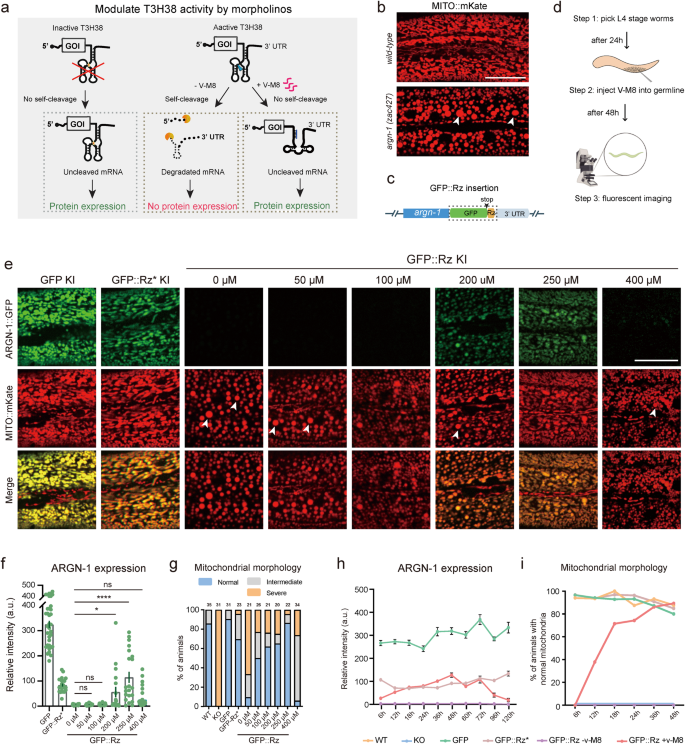
After demonstrating that T3H38 ribozyme may very well be used to control endogenous gene expression from “On” to “Off”, we sought to find out whether or not it might probably act as an “Off-to-On” device. Antisense oligonucleotide v-M8 induces a conformational change of T3H38, thereby blocking the self-cleavage exercise of T3H38 ribozyme10. This technique efficiently induced the expression of an exogenous transgene in mice (Fig. 5a). To check this, we chosen argn-1 because the goal, utilizing strains carrying gfp, gfp::Rz* or gfp::Rz insertion at argn-1 locus (Fig. 5b, c). We then first tried whether or not injecting the v-M8 morpholino might regulate the expression of endogenous genes from “Off” to “On” state. We injected v-M8 into the argn-1::gfp::Rz animals on day 1 of maturity and detected the protein expression after 48 h (Fig. 5d). Quantitative evaluation of ARGN-1::GFP ranges revealed that endogenous argn-1 expression was partially recovered in a dose-dependent method. Injection of morpholino at 250 μM efficiently restored the gene expression to ranges corresponding to these of the inactive T3H38 management group, suggesting an environment friendly block of the ribozyme-mediated self-cleavage (Fig. 5e, f). We additionally examined the mitochondrial morphology within the animals injected with morpholino. Roughly 50% to 80% of animals confirmed wild-type-like regular mitochondrial morphology when injected with 50 to 250 μM morpholino. Furthermore, 250 μM morpholino confirmed the very best efficiency, which is according to the expression stage (Fig. 5g). Notably, the injection of 400 μM morpholino was ineffective in turning on the expression of endogenous ARGN-1 or rescuing the irregular mitochondrial morphology. The underlying mechanism behind this commentary requires future investigation (Fig. 5e–g). Comparable restoration of endogenous expression and protein perform by injecting antisense morpholino was noticed for scav-3 and sax-7 (Supplementary Figs. S4a–c and S5a, b).
a Diagram representing how an antisense oligonucleotide inactivates a ribozyme to revive protein expression and relative controls. Tailored with permission from Wurmthaler et al.11. b Confocal photos to point out the morphology of mitochondria in wild-type and mutant animals. Scale bars: 20 μm. c Diagram displaying the cassette (outlined by a field) built-in into the C-terminal of argn-1. d A workflow of injecting morpholinos into the worm germ line and confocal imaging for quantifications. e Fluorescence photos to point out the morphology of mitochondrial and ARGN-1 expression underneath varied circumstances. Scale bar, 20 μm. f Relative protein expression of ARGN-1::GFP underneath completely different circumstances as proven in (e). Every dot represents a single worm. Variety of animals quantified: n = 35 for ARGN-1::GFP; n = 24 for ARGN-1::GFP::Rz*; n = 19 for ARGN-1::GFP::Rz with out v-M8; n = 27 for ARGN-1::GFP::Rz injected with 50 μM v-M8; n = 21 for ARGN-1::GFP::Rz injected with 100 μM v-M8; n = 18 for ARGN-1::GFP::Rz injected with 200 μM v-M8; n = 21 for ARGN-1::GFP::Rz injected with 250 μM v-M8; n = 39 for ARGN-1::GFP::Rz injected with 400 μM v-M8. All information have been proven as imply ± s.e.m. Ns: not vital. *p < 0.05. ****p < 0.0001 (one-way ANOVA with the Tukey correction). g Quantifications of mitochondria morphology underneath completely different circumstances as proven in (e). The variety of animals quantified for every group was indicated above every column. h The expression of ARGN-1::GFP was measured primarily based on fluorescent imaging at indicated time factors (hours after V-M8 morpholino or M9 injection) for varied genetic backgrounds (GFP, GFP::Rz* or GFP::Rz). All values are displayed as imply ± s.e.m. n ≥ 19 animals. The precise variety of animals is offered as supply information. i The proportion of GFP-Rz animals containing regular mitochondrial was quantified at indicated time factors after V-M8 morpholino or M9 injection. n ≥ 26 animals. The precise variety of animals is offered as supply information. Arrows in (b) and (e): enlarged mitochondria. Supply information are offered as a Supply Information file.
To research the kinetics of the endogenous gene expression induced by morpholino, we carried out a time-course evaluation of protein expression from the argn-1::gfp::energetic T3H38 locus put up morpholino injections. A gentle induction was noticed at 6 h. The expression reached the very best stage at 48 h after which began to say no. At 120 h post-injection, the expression stage was virtually undetectable (Fig. 5h). We then analyzed mitochondria morphology to find out the purposeful final result of endogenous gene expression. At 6 h post-injection, not one of the animals confirmed regular mitochondrial morphology, suggesting that the gentle expression stage is inadequate for the ARGN-1’s perform. The proportion of animals displaying regular mitochondrial morphology elevated dramatically between 12 and 36 h and peaked at 48 h after injection, which was corresponding to the inactive T3H38 management group (Fig. 5i). Comparable induction was noticed for endogenous scav-3 by injection of morpholino (Supplementary Fig. S4a–c).
Cre/loxP-mediated Ribo-On activates gene expression in area and time
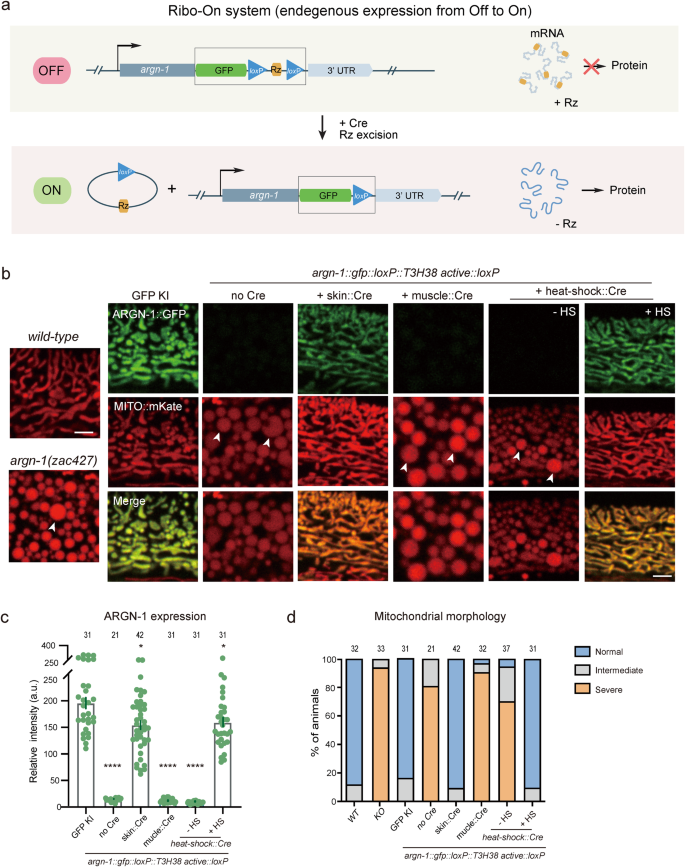
The morpholino injection technique can manipulate the expression of endogenous genes with temporal decision however lacks spatial regulation. As well as, our findings counsel that the incorporation of an inactive ribozyme can perturb the expression of some endogenous genes we examined, probably making it difficult to completely restore their expression ranges to that of wild-type animals through morpholino injection. Subsequently, impressed by the ribozyme-Cre-loxP-dependent conditional Ribo-Off technique, we inserted the coding sequence for gfp::loxP::Rz::loxP instantly after the cease codons of endogenous genes. Theoretically, inserting the energetic ribozyme between the coding area and the three’ UTR would trigger mRNA degradation and abolish the gene expression. Utilizing the Cre recombinase, the expression may very well be restored in both a spatial or a temporal method (Fig. 6a). To check the thought, the gfp::loxP::Rz::loxP cassette was built-in instantly after the cease codon of argn-1 gene, which did generate enlarged mitochondria just like that of loss-of-function mutants. Pores and skin-expressed Cre, however not the muscle-expressed Cre, restored each the mitochondria morphology and epidermal expression, suggesting that argn-1 was efficiently restored particularly within the dermis. Utilizing heat-shock-induced Cre, we have been in a position to induce ARGN-1::GFP expression and rescue the argn-1 loss-of-function phenotype in a temporal method (Fig. 6b–d). Comparable Off-to-On regulation was achieved for dma-122 (Supplementary Fig. S6a, b). We subsequently named this ribozyme-dependent expression ON-switch as Ribo-On. Collectively, our outcomes counsel that Ribo-On and Ribo-Off are efficient instruments for manipulating endogenous gene expression each in area and time, thus facilitating purposeful research of particular genes in vivo.
a Schematic representing how ribozyme/Cre-loxp methods conditionally activate gene expression. b The expression of ARGN-1::GFP within the dermis and the mitochondrial morphology in one-day-old grownup worms underneath completely different genetic backgrounds. Cre expression was spatially managed by tissue-specific promoters or temporally induced by heat-shock promoter, respectively. Scale bars: 5 μm. Arrows: enlarged mitochondria. c Quantification of ARGN-1::GFP expression within the dermis underneath completely different circumstances, as proven in (b). Every dot represents a single worm, n ≥ 25. The precise variety of animals is offered as supply information. All information are introduced as imply ± s.e.m. *p < 0.05. ****p < 0.0001 (one-way ANOVA with the Tukey correction). d Quantifications of the proportion of animals containing regular, intermediate faulty or extreme faulty morphology of mitochondrial within the dermis underneath completely different circumstances as proven in (b). The variety of animals quantified was proven above every column. Supply information are offered as a Supply Information file.